PDF (Chapter 7)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Use of Mathematical Modeling and Other Biophysical Methods For
USE OF MATHEMATICAL MODELING AND OTHER BIOPHYSICAL METHODS FOR INSIGHTS INTO IRON-RELATED PHENOMENA OF BIOLOGICAL SYSTEMS A Dissertation by JOSHUA D. WOFFORD Submitted to the Office of Graduate and Professional Studies of Texas A&M University in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY Chair of Committee, Paul A. Lindahl Committee Members, David P. Barondeau Simon W. North Vishal M. Gohil Head of Department, Simon W. North December 2018 Major Subject: Chemistry Copyright 2018 Joshua D. Wofford ABSTRACT Iron is a crucial nutrient in most living systems. It forms the active centers of many proteins that are critical for many cellular functions, either by themselves or as Fe-S clusters and hemes. However, Fe is potentially toxic to the cell in high concentrations and must be tightly regulated. There has been much work into understanding various pieces of Fe trafficking and regulation, but integrating all of this information into a coherent model has proven difficult. Past research has focused on different Fe species, including cytosolic labile Fe or mitochondrial Fe-S clusters, as being the main regulator of Fe trafficking in yeast. Our initial modeling efforts demonstrate that both cytosolic Fe and mitochondrial ISC assembly are required for proper regulation. More recent modeling efforts involved a more rigorous multi- tiered approach. Model simulations were optimized against experimental results involving respiring wild-type and Mrs3/4-deleted yeast. Simulations from both modeling studies suggest that mitochondria possess a “respiratory shield” that prevents a vicious cycle of nanoparticle formation, ISC loss, and subsequent loading of mitochondria with iron. -
Nitroxide-Mediated Polymerization
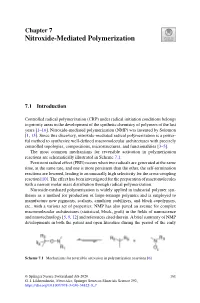
Chapter 7 Nitroxide-Mediated Polymerization 7.1 Introduction Controlled radical polymerization (CRP) under radical initiation conditions belongs to priority areas in the development of the synthetic chemistry of polymers of the last years [1–16]. Nitroxide-mediated polymerization (NMP) was invented by Solomon [1, 13]. Since this discovery, nitroxide-mediated radical polymerization is a power- ful method to synthesize well-defined macromolecular architectures with precisely controlled topologies, compositions, microstructures, and functionalities [3–5]. The most common mechanisms for reversible activation in polymerization reactions are schematically illustrated in Scheme 7.1. Persistent radical effect (PRE) occurs when two radicals are generated at the same time, at the same rate, and one is more persistent than the other, the self-termination reactions are lowered, leading to an unusually high selectivity for the cross-coupling reaction [10]. The effect has been investigated for the preparation of macromolecules with a narrow molar mass distribution through radical polymerization. Nitroxide-mediated polymerization is widely applied in industrial polymer syn- theses as a method for production of large-tonnage polymers and is employed to manufacture new pigments, sealants, emulsion stabilizers, and block copolymers, etc., with a various set of properties. NMP has also paved an avenue for complex macromolecular architectures (statistical, block, graft) in the fields of nanoscience and nanotechnology [5, 9, 12] and references cited therein. A brief summary of NMP developments in both the patent and open literature during the period of the early Scheme 7.1 Mechanisms for reversible activation in polymerization reactions [6] © Springer Nature Switzerland AG 2020 161 G. I. Likhtenshtein, Nitroxides, Springer Series in Materials Science 292, https://doi.org/10.1007/978-3-030-34822-9_7 162 7 Nitroxide-Mediated Polymerization 1980–2000 was presented in [11]. -

Electron Flow and Management in Living Systems: Advancing Understanding of Electron Transfer to Nitrogenase
Utah State University DigitalCommons@USU All Graduate Theses and Dissertations Graduate Studies 8-2018 Electron Flow and Management in Living Systems: Advancing Understanding of Electron Transfer to Nitrogenase Rhesa N. Ledbetter Utah State University Follow this and additional works at: https://digitalcommons.usu.edu/etd Part of the Biochemistry Commons Recommended Citation Ledbetter, Rhesa N., "Electron Flow and Management in Living Systems: Advancing Understanding of Electron Transfer to Nitrogenase" (2018). All Graduate Theses and Dissertations. 7197. https://digitalcommons.usu.edu/etd/7197 This Dissertation is brought to you for free and open access by the Graduate Studies at DigitalCommons@USU. It has been accepted for inclusion in All Graduate Theses and Dissertations by an authorized administrator of DigitalCommons@USU. For more information, please contact [email protected]. ELECTRON FLOW AND MANAGEMENT IN LIVING SYSTEMS: ADVANCING UNDERSTANDING OF ELECTRON TRANSFER TO NITROGENASE by Rhesa N. Ledbetter A dissertation submitted in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY in Biochemistry Approved: ______________________ ______________________ Lance C. Seefeldt, Ph.D. Scott A. Ensign, Ph.D. Biochemistry Biochemistry Major Professor Committee Member ______________________ ______________________ Bruce Bugbee, Ph.D. Sean J. Johnson, Ph.D. Plant Physiology Biochemistry Committee Member Committee Member ______________________ ______________________ Nicholas E. Dickenson, Ph.D. Mark R. McLellan, Ph.D. Biochemistry Vice President for Research and Committee Member Dean of the School of Graduate Studies UTAH STATE UNIVERSITY Logan, Utah 2018 ii Copyright © Rhesa N. Ledbetter 2018 All Rights Reserved iii ABSTRACT Electron Flow and Management in Living Systems: Advancing Understanding of Electron Transfer to Nitrogenase by Rhesa N. -

Electrochemical and Structural Characterization of Azotobacter Vinelandii Flavodoxin II
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Caltech Authors Electrochemical and structural characterization of Azotobacter vinelandii flavodoxin II Helen M. Segal,1 Thomas Spatzal,1 Michael G. Hill,2 Andrew K. Udit,2 and Douglas C. Rees 1* 1Division of Chemistry and Chemical Engineering, Howard Hughes Medical Institute, California Institute of Technology, Pasadena, California 91125 2Division of Chemistry, Occidental College, Los Angeles, California 90041 Received 1 June 2017; Accepted 10 July 2017 DOI: 10.1002/pro.3236 Published online 14 July 2017 proteinscience.org Abstract: Azotobacter vinelandii flavodoxin II serves as a physiological reductant of nitrogenase, the enzyme system mediating biological nitrogen fixation. Wildtype A. vinelandii flavodoxin II was electrochemically and crystallographically characterized to better understand the molecular basis for this functional role. The redox properties were monitored on surfactant-modified basal plane graphite electrodes, with two distinct redox couples measured by cyclic voltammetry correspond- ing to reduction potentials of 2483 6 1 mV and 2187 6 9 mV (vs. NHE) in 50 mM potassium phos- phate, 150 mM NaCl, pH 7.5. These redox potentials were assigned as the semiquinone/ hydroquinone couple and the quinone/semiquinone couple, respectively. This study constitutes one of the first applications of surfactant-modified basal plane graphite electrodes to characterize the redox properties of a flavodoxin, thus providing a novel electrochemical method to study this class of protein. The X-ray crystal structure of the flavodoxin purified from A. vinelandii was solved at 1.17 A˚ resolution. With this structure, the native nitrogenase electron transfer proteins have all been structurally characterized. -

Cluster Characterization in Iron-Sulfur Proteins by Magnetic Circular Dichroism (Spectroscopic Probes/Ferredoxins) P
Proc. Natl. Acad. Sci. USA Vol. 75, No. 11, pp. 5273-5275, November 1978 Biochemistry Cluster characterization in iron-sulfur proteins by magnetic circular dichroism (spectroscopic probes/ferredoxins) P. J. STEPHENS*, A. J. THOMSON*t, T. A. KEIDERLING*t, J. RAWLINGS*§, K. K. RAOT, AND D. 0. HALLS * Department of Chemistry, University of Southern California, Los Angeles, California 90007; and ISchool of Biological Sciences, University of London King's College, 68 Half Moon Lane, London, England Communicated by Martin D. Kamen, August 2,1978- ABSTRACT We report magnetic circular dichroism (MCD) respect to the number of 4-Fe clusters). Ac values are normal- spectra of 4-Fe iron-sulfur clusters in the iron-sulfur proteins ized to a magnetic field of 10 kilogauss. Chromatium high-potential iron protein (HIPIP), Bacillus 1-3 MCD and stearothernophilus ferredoxin and Clostridium pasteurianum Figs. display absorption spectra for clusters ferredoxin. The MCD is found to vary significantly with cluster in the C2-, C3-, and Cl- states, respectively. The absorption oxidation state but is relatively insensitive to the nature of the spectra are typical of 4-Fe clusters, exhibiting few distinct protein. The spectra obtained are compared with the corre- features"l; for a given oxidation state the spectra are insensitive sponding spectra of iron-sulfur proteins containing 2-Fe clus- to the specific protein under study. By comparison, the MCD ters. It is concluded that MCD is useful for the characterization spectra are appreciably more structured than the absorption of iron-sulfur cluster type and oxidation state in iron-sulfur spectra but retain the insensitivity to the nature of the proteins and is superior for this purpose to absorption and nat- associated ural circular dichroism spectroscopy. -

Electron Transfer Partners of Cytochrome P450
4 Electron Transfer Partners of Cytochrome P450 Mark J.l. Paine, Nigel S. Scrutton, Andrew W. Munro, Aldo Gutierrez, Gordon C.K. Roberts, and C. Roland Wolf 1. Introduction Although P450 redox partners are usually expressed independently, "self-sufficient" P450 monooxygenase systems have also evolved through Cytochromes P450 contain a heme center the fusion of P450 and CPR genes. These fusion where the activation of molecular oxygen occurs, molecules are found in bacteria and fungi, the best- resulting in the insertion of a single atom of known example being P450 BM3, a fatty acid oxygen into an organic substrate with the con (0-2 hydroxylase from Bacillus megaterium, which comitant reduction of the other atom to water. The comprises a soluble P450 with a fiised carboxyl- monooxygenation reaction requires a coupled and terminal CPR module (recently reviewed by stepwise supply of electrons, which are derived Munro^). BM3 has the highest catalytic activity from NAD(P)H and supplied via a redox partner. known for a P450 monooxygenase^ and was for P450s are generally divided into two major classes many years the only naturally occurring ftised sys (Class I and Class II) according to the different tem known until the identification of a eukaryotic types of electron transfer systems they use. P450s membrane-bound equivalent fatty acid hydroxy in the Class I family include bacterial and mito lase, CYP505A1, from the phytopathogenic fungus chondrial P450s, which use a two-component Fusarium oxysporurrP. A number of novel P450 sys shuttle system consisting of an iron-sulfur protein tems are starting to emerge from the large numbers (ferredoxin) and ferredoxin reductase (Figure 4.1). -

Containing Aldehyde Ferredoxin Oxidoreductase From
JOURNAL OF BACTERIOLOGY, Aug. 1995, p. 4817–4819 Vol. 177, No. 16 0021-9193/95/$04.0010 Copyright q 1995, American Society for Microbiology Molecular Characterization of the Genes Encoding the Tungsten- Containing Aldehyde Ferredoxin Oxidoreductase from Pyrococcus furiosus and Formaldehyde Ferredoxin Oxidoreductase from Thermococcus litoralis ARNULF KLETZIN,1† SWARNALATHA MUKUND,1 TERRY L. KELLEY-CROUSE,1 2 2 1 MICHAEL K. CHAN, DOUGLAS C. REES, AND MICHAEL W. W. ADAMS * Department of Biochemistry and Molecular Biology and Center for Metalloenzyme Studies, University of Georgia, Athens, Georgia 30602,1 and Division of Chemistry, California Institute of Technology, Pasadena, California 911252 Received 21 February 1995/Accepted 8 June 1995 The hyperthermophilic archaea Pyrococcus furiosus and Thermococcus litoralis contain the tungstoenzymes aldehyde ferredoxin oxidoreductase, a homodimer, and formaldehyde ferredoxin oxidoreductase, a homotet- ramer. Herein we report the cloning and sequencing of the P. furiosus gene aor (605 residues; Mr, 66,630) and the T. litoralis gene for (621 residues; Mr, 68,941). Enzymes containing tungsten (W) are rare in biology, yet the first 169 amino acid residues of a P. furiosus ahc gene encoding hyperthermophilic archaea contain three distinct types, all of an S-adenosylhomocysteine hydrolase; and (iii) a short open which catalyze aldehyde oxidation (6–9). The homodimeric reading frame and a partially sequenced long open reading aldehyde ferredoxin oxidoreductase (AOR) of Pyrococcus fu- frame of unknown function (Fig. 1) (5). riosus (maximum growth temperature [Tmax], 1058C [3]) oxi- The gene for AOR contained 605 codons which correspond dizes a wide range of aliphatic and aromatic, nonphosphory- to a protein with a molecular weight of 66,630 (compared with lated aldehydes (7). -

Glycolysis Citric Acid Cycle Oxidative Phosphorylation Calvin Cycle Light
Stage 3: RuBP regeneration Glycolysis Ribulose 5- Light-Dependent Reaction (Cytosol) phosphate 3 ATP + C6H12O6 + 2 NAD + 2 ADP + 2 Pi 3 ADP + 3 Pi + + 1 GA3P 6 NADP + H Pi NADPH + ADP + Pi ATP 2 C3H4O3 + 2 NADH + 2 H + 2 ATP + 2 H2O 3 CO2 Stage 1: ATP investment ½ glucose + + Glucose 2 H2O 4H + O2 2H Ferredoxin ATP Glyceraldehyde 3- Ribulose 1,5- Light Light Fx iron-sulfur Sakai-Kawada, F Hexokinase phosphate bisphosphate - 4e + center 2016 ADP Calvin Cycle 2H Stroma Mn-Ca cluster + 6 NADP + Light-Independent Reaction Phylloquinone Glucose 6-phosphate + 6 H + 6 Pi Thylakoid Tyr (Stroma) z Fe-S Cyt f Stage 1: carbon membrane Phosphoglucose 6 NADPH P680 P680* PQH fixation 2 Plastocyanin P700 P700* D-(+)-Glucose isomerase Cyt b6 1,3- Pheophytin PQA PQB Fructose 6-phosphate Bisphosphoglycerate ATP Lumen Phosphofructokinase-1 3-Phosphoglycerate ADP Photosystem II P680 2H+ Photosystem I P700 Stage 2: 3-PGA Photosynthesis Fructose 1,6-bisphosphate reduction 2H+ 6 ADP 6 ATP 6 CO2 + 6 H2O C6H12O6 + 6 O2 H+ + 6 Pi Cytochrome b6f Aldolase Plastoquinol-plastocyanin ATP synthase NADH reductase Triose phosphate + + + CO2 + H NAD + CoA-SH isomerase α-Ketoglutarate + Stage 2: 6-carbonTwo 3- NAD+ NADH + H + CO2 Glyceraldehyde 3-phosphate Dihydroxyacetone phosphate carbons Isocitrate α-Ketoglutarate dehydogenase dehydrogenase Glyceraldehyde + Pi + NAD Isocitrate complex 3-phosphate Succinyl CoA Oxidative Phosphorylation dehydrogenase NADH + H+ Electron Transport Chain GDP + Pi 1,3-Bisphosphoglycerate H+ Succinyl CoA GTP + CoA-SH Aconitase synthetase -

Energy Conservation Involving 2 Respiratory Circuits
Energy conservation involving 2 respiratory circuits Marie Charlotte Schoelmericha,1 , Alexander Katsyva, Judith Donig¨ a, Timothy J. Hackmannb, and Volker Muller¨ a,2 aMolecular Microbiology and Bioenergetics, Institute of Molecular Biosciences, Johann Wolfgang Goethe University Frankfurt/Main, 60438 Frankfurt, Germany; and bDepartment of Animal Science, University of California, Davis, CA 95616 Edited by Caroline S. Harwood, University of Washington, Seattle, WA, and approved November 27, 2019 (received for review August 28, 2019) Chemiosmosis and substrate-level phosphorylation are the 2 acetogenic bacteria. Acetogens use the reductive acetyl- mechanisms employed to form the biological energy currency coenzyme A (acetyl-CoA) pathway to fix CO2 using inorganic adenosine triphosphate (ATP). During chemiosmosis, a transmem- gases such as H2 or CO (autotrophic) or organic compounds brane electrochemical ion gradient is harnessed by a rotary ATP such as sugars (heterotrophic) as an electron source. Under synthase to phosphorylate adenosine diphosphate to ATP. In autotrophic conditions, they rely on a chemiosmotic mechanism microorganisms, this ion gradient is usually composed of H+, to conserve energy in the form of ATP. Ferredoxin (Fd) is the but it can also be composed of Na+. Here, we show that the central electron carrier in bioenergetics of acetogens and fuels strictly anaerobic rumen bacterium Pseudobutyrivibrio ruminis 2 distinct respiratory enzymes, the Fd2−:NAD+ oxidoreductase possesses 2 ATP synthases and 2 distinct respiratory enzymes, (Rnf complex) and the Fd2−:H+ oxidoreductase (Ech complex) the ferredoxin:NAD+ oxidoreductase (Rnf complex) and the (2–4). The Rnf complex in Acetobacterium woodii establishes a energy-converting hydrogenase (Ech complex). In silico analyses Na+ gradient, which fuels a Na+-dependent ATP synthase. -

Electron Carriers: Proteins and Cofactors in Oxidative Phosphorylation
Electron Carriers: Proteins Secondary article and Cofactors in Oxidative Article Contents . Introduction . Overview of Oxidative Phosphorylation and Phosphorylation Photophosphorylation . Electron Carriers: NADH, Flavoproteins, Cytochromes, Klaus-Heinrich Roehm, Philipps University, Marburg, Germany Iron–Sulfur Proteins, Quinones and Others . Reduction Potentials Biological electron transfer is mediated by a heterogeneous group of redox cofactors. Most . Electron Transfer Mechanisms of these compounds are firmly bound by proteins, others migrate freely within membranes, and some are soluble. The orderly flow of electrons through chains of such cofactors is fundamental to the generation of metabolic energy. Introduction schematic way, the general topology of both chains and the Most of the metabolic energy generated in living cells approximate size of the individual complexes as deter- results from processes that abstract electrons from a donor mined by X-ray crystallography or electron microscopy. molecule, channel them through an electron transport The types and numbers of the cofactor molecules chain, and finally deliver them to an acceptor molecule. participating in electron transport are also indicated. Their When the donor is NADH and the acceptor is oxygen, the chemical properties are discussed in more detail below. overall reaction is strongly exergonic and ATPcan be generated (oxidative phosphorylation). In the light reac- tion of photosynthesis, electrons are taken from water and The respiratory chain 1 eventually transferred to NADP to yield NADPH. This Primary electron donors of respiratory electron transport is an endergonic process, and therefore has to be driven by in the mitochondria are either NADH 1 H 1 or metabo- the absorption of light energy. The resulting overall lites that can be oxidized by FAD-dependent dehydro- reaction becomes sufficiently exergonic to yield significant genases (Figure 1a). -

Consequences of the Structure of the Cytochrome B6 F Complex for Its Charge Transfer Pathways ⁎ William A
Biochimica et Biophysica Acta 1757 (2006) 339–345 http://www.elsevier.com/locate/bba Review Consequences of the structure of the cytochrome b6 f complex for its charge transfer pathways ⁎ William A. Cramer , Huamin Zhang Department of Biological Sciences, Purdue University, West Lafayette, IN 47907, USA Received 17 January 2006; received in revised form 30 March 2006; accepted 24 April 2006 Available online 4 May 2006 Abstract At least two features of the crystal structures of the cytochrome b6 f complex from the thermophilic cyanobacterium, Mastigocladus laminosus and a green alga, Chlamydomonas reinhardtii, have implications for the pathways and mechanism of charge (electron/proton) transfer in the complex: (i) The narrow 11×12 Å portal between the p-side of the quinone exchange cavity and p-side plastoquinone/quinol binding niche, through which all Q/QH2 must pass, is smaller in the b6 f than in the bc1 complex because of its partial occlusion by the phytyl chain of the one bound chlorophyll a molecule in the b6 f complex. Thus, the pathway for trans-membrane passage of the lipophilic quinone is even more labyrinthine in the b6 f than in the bc1 complex. (ii) A unique covalently bound heme, heme cn, in close proximity to the n-side b heme, is present in the b6 f complex. The b6 f structure implies that a Q cycle mechanism must be modified to include heme cn as an intermediate between heme bn and plastoquinone bound at a different site than in the bc1 complex. In addition, it is likely that the heme bn–cn couple participates in photosytem + I-linked cyclic electron transport that requires ferredoxin and the ferredoxin: NADP reductase. -

Bacterial Ferredoxin' R
BACTERIOLOGICAL REVIEWS Vol. 28, No. 4, p. 497-517 December, 1964 Copyright @ 1964 American Society for Microbiology Printed in U.S.A. BACTERIAL FERREDOXIN' R. C. VALENTINE Department of Biochemistry, University of California, Berkeley, California INTRODUCTION ................................................... 497 Low-Potential Electron Transport................................................... 498 ISOLATION OF FERREDOXIN FROM BACTERIA ............................................... 500 Purification ................................................... 500 Assays................................................... 500 Phosphoroclastic Assay ................. 501 Hydrogenase Assay ................ 502 Other Assays .................... 502 Distribution................ 502 PROPERTIES OF CRYSTALLINE FERREDOXIN ................ 503 Oxidized and Reduced States ................ 503 Spectra ................ 504 Composition ......................................................... 504 ENZYMATIC REDUCTION OF FERREDOXIN ................................................... 505 Phosphoroclastic System ................................................... 505 a-Ketoglutarate ................................................... 506 Dithionite as H2 Precursor ................................................... 507 Hypoxanthine ................................................... 507 Coupling with Formate................................................... 507 OXIDATION OF REDUCED FERREDOXIN ................................................... 507 Urate ..................................................