A Mathematical Model of Iron Import and Trafficking in Wild-Type and Mrs3/4ΔΔ Yeast Cells Joshua D
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Use of Mathematical Modeling and Other Biophysical Methods For
USE OF MATHEMATICAL MODELING AND OTHER BIOPHYSICAL METHODS FOR INSIGHTS INTO IRON-RELATED PHENOMENA OF BIOLOGICAL SYSTEMS A Dissertation by JOSHUA D. WOFFORD Submitted to the Office of Graduate and Professional Studies of Texas A&M University in partial fulfillment of the requirements for the degree of DOCTOR OF PHILOSOPHY Chair of Committee, Paul A. Lindahl Committee Members, David P. Barondeau Simon W. North Vishal M. Gohil Head of Department, Simon W. North December 2018 Major Subject: Chemistry Copyright 2018 Joshua D. Wofford ABSTRACT Iron is a crucial nutrient in most living systems. It forms the active centers of many proteins that are critical for many cellular functions, either by themselves or as Fe-S clusters and hemes. However, Fe is potentially toxic to the cell in high concentrations and must be tightly regulated. There has been much work into understanding various pieces of Fe trafficking and regulation, but integrating all of this information into a coherent model has proven difficult. Past research has focused on different Fe species, including cytosolic labile Fe or mitochondrial Fe-S clusters, as being the main regulator of Fe trafficking in yeast. Our initial modeling efforts demonstrate that both cytosolic Fe and mitochondrial ISC assembly are required for proper regulation. More recent modeling efforts involved a more rigorous multi- tiered approach. Model simulations were optimized against experimental results involving respiring wild-type and Mrs3/4-deleted yeast. Simulations from both modeling studies suggest that mitochondria possess a “respiratory shield” that prevents a vicious cycle of nanoparticle formation, ISC loss, and subsequent loading of mitochondria with iron. -
Nitroxide-Mediated Polymerization
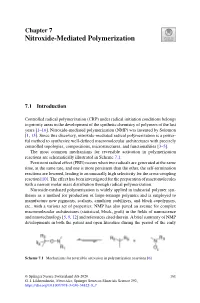
Chapter 7 Nitroxide-Mediated Polymerization 7.1 Introduction Controlled radical polymerization (CRP) under radical initiation conditions belongs to priority areas in the development of the synthetic chemistry of polymers of the last years [1–16]. Nitroxide-mediated polymerization (NMP) was invented by Solomon [1, 13]. Since this discovery, nitroxide-mediated radical polymerization is a power- ful method to synthesize well-defined macromolecular architectures with precisely controlled topologies, compositions, microstructures, and functionalities [3–5]. The most common mechanisms for reversible activation in polymerization reactions are schematically illustrated in Scheme 7.1. Persistent radical effect (PRE) occurs when two radicals are generated at the same time, at the same rate, and one is more persistent than the other, the self-termination reactions are lowered, leading to an unusually high selectivity for the cross-coupling reaction [10]. The effect has been investigated for the preparation of macromolecules with a narrow molar mass distribution through radical polymerization. Nitroxide-mediated polymerization is widely applied in industrial polymer syn- theses as a method for production of large-tonnage polymers and is employed to manufacture new pigments, sealants, emulsion stabilizers, and block copolymers, etc., with a various set of properties. NMP has also paved an avenue for complex macromolecular architectures (statistical, block, graft) in the fields of nanoscience and nanotechnology [5, 9, 12] and references cited therein. A brief summary of NMP developments in both the patent and open literature during the period of the early Scheme 7.1 Mechanisms for reversible activation in polymerization reactions [6] © Springer Nature Switzerland AG 2020 161 G. I. Likhtenshtein, Nitroxides, Springer Series in Materials Science 292, https://doi.org/10.1007/978-3-030-34822-9_7 162 7 Nitroxide-Mediated Polymerization 1980–2000 was presented in [11]. -

Studies of Iron Sulfur Cluster Maturation and Transport DISSERTATION Presented in Partial Fulfillment of the Requirements for Th
Studies of Iron Sulfur Cluster Maturation and Transport DISSERTATION Presented in Partial Fulfillment of the Requirements for the Degree Doctor of Philosophy in the Graduate School of The Ohio State University By Jingwei Li Graduate Program in Chemistry The Ohio State University 2015 Dissertation Committee: Professor James A. Cowan, Advisor Professor Ross E. Dalbey Professor Claudia Turro Copyright by Jingwei Li 2015 Abstract Cellular iron homeostasis is critically dependent on sensory and regulatory mechanisms that maintain a balance of intracellular iron concentrations. Divergence from a healthy iron concentration can result in common disease states such as anemia and ataxia. With the goal of understanding the molecular basis for such health problems, and advancing the knowledge based toward potential remedies, an understanding of the molecular details of cellular iron transport and the biological chemistry of iron species is an essential prerequisite. In that regard, iron-sulfur clusters are ubiquitous iron-containing centers in a variety of proteins and serve a multitude of roles that include electron transfer, catalysis of reactions, and sensors of cellular oxygen and iron levels. Recently, a substantial body of evidence has suggested an essential role for cellular glutathione (a molecule normally implicated with eliminating reactive oxygen species from cells) in the regulation, stabilization and biosynthesis of cellular iron-sulfur clusters in humans and other complex organisms. We have demonstrated that glutathione can naturally bind to iron-sulfur cluster precursors and have isolated and characterized this species and shown it to be stable under physiological conditions. More importantly, we have demonstrated that the glutathione-bound iron-sulfur cluster can be transported by a ii critical export protein from the cellular mitochondrion. -

Electrochemical and Structural Characterization of Azotobacter Vinelandii Flavodoxin II
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Caltech Authors Electrochemical and structural characterization of Azotobacter vinelandii flavodoxin II Helen M. Segal,1 Thomas Spatzal,1 Michael G. Hill,2 Andrew K. Udit,2 and Douglas C. Rees 1* 1Division of Chemistry and Chemical Engineering, Howard Hughes Medical Institute, California Institute of Technology, Pasadena, California 91125 2Division of Chemistry, Occidental College, Los Angeles, California 90041 Received 1 June 2017; Accepted 10 July 2017 DOI: 10.1002/pro.3236 Published online 14 July 2017 proteinscience.org Abstract: Azotobacter vinelandii flavodoxin II serves as a physiological reductant of nitrogenase, the enzyme system mediating biological nitrogen fixation. Wildtype A. vinelandii flavodoxin II was electrochemically and crystallographically characterized to better understand the molecular basis for this functional role. The redox properties were monitored on surfactant-modified basal plane graphite electrodes, with two distinct redox couples measured by cyclic voltammetry correspond- ing to reduction potentials of 2483 6 1 mV and 2187 6 9 mV (vs. NHE) in 50 mM potassium phos- phate, 150 mM NaCl, pH 7.5. These redox potentials were assigned as the semiquinone/ hydroquinone couple and the quinone/semiquinone couple, respectively. This study constitutes one of the first applications of surfactant-modified basal plane graphite electrodes to characterize the redox properties of a flavodoxin, thus providing a novel electrochemical method to study this class of protein. The X-ray crystal structure of the flavodoxin purified from A. vinelandii was solved at 1.17 A˚ resolution. With this structure, the native nitrogenase electron transfer proteins have all been structurally characterized. -

Role of Rim101p in the Ph Response in Candida Albicans Michael Weyler
Role of Rim101p in the pH response in Candida albicans Michael Weyler To cite this version: Michael Weyler. Role of Rim101p in the pH response in Candida albicans. Biomolecules [q-bio.BM]. Université Paris Sud - Paris XI, 2007. English. tel-00165802 HAL Id: tel-00165802 https://tel.archives-ouvertes.fr/tel-00165802 Submitted on 27 Jul 2007 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. UNIVERSITE PARISXI UFR SCIENTIFIQUE D’ORSAY THESE présentée par Michael Weyler pour obtenir le grade de DOCTEUR EN SCIENCES DE L’UNIVERSITE PARISXI-ORSAY LE RÔLE DE RIM101p DANS LA RÉPONSE AU pH CHEZ CANDIDA ALBICANS Soutenance prévue le 6 juillet 2007 devant le jury composé de: Pr. Dr. H. Delacroix Président Dr. J-M. Camadro Rapporteur Pr. Dr. F. M. Klis Rapporteur Dr. G. Janbon Examinateur Dr. M. Lavie-Richard Examinateur Pr. Dr. C. Gaillardin Examinateur Remerciements Tout d’abord je voudrais remercier vivement mon directeur de thèse, Prof. Claude Gaillardin, pour m’avoir permis d’effectuer ce travail au sein de son laboratoire, pour ses conseils et sa disponibilité malgré son calendrier bien remplis. Je lui remercie également pour m’avoir laissé beaucoup de liberté dans mon travail, et pour la possibilité de participer aux différents congrès au cours de ma formation de thèse. -

Programmed Cell-Death by Ferroptosis: Antioxidants As Mitigators
International Journal of Molecular Sciences Review Programmed Cell-Death by Ferroptosis: Antioxidants as Mitigators Naroa Kajarabille 1 and Gladys O. Latunde-Dada 2,* 1 Nutrition and Obesity Group, Department of Nutrition and Food Sciences, University of the Basque Country (UPV/EHU), 01006 Vitoria, Spain; [email protected] 2 King’s College London, Department of Nutritional Sciences, Faculty of Life Sciences and Medicine, Franklin-Wilkins Building, 150 Stamford Street, London SE1 9NH, UK * Correspondence: [email protected] Received: 9 September 2019; Accepted: 2 October 2019; Published: 8 October 2019 Abstract: Iron, the fourth most abundant element in the Earth’s crust, is vital in living organisms because of its diverse ligand-binding and electron-transfer properties. This ability of iron in the redox cycle as a ferrous ion enables it to react with H2O2, in the Fenton reaction, to produce a hydroxyl radical ( OH)—one of the reactive oxygen species (ROS) that cause deleterious oxidative damage • to DNA, proteins, and membrane lipids. Ferroptosis is a non-apoptotic regulated cell death that is dependent on iron and reactive oxygen species (ROS) and is characterized by lipid peroxidation. It is triggered when the endogenous antioxidant status of the cell is compromised, leading to lipid ROS accumulation that is toxic and damaging to the membrane structure. Consequently, oxidative stress and the antioxidant levels of the cells are important modulators of lipid peroxidation that induce this novel form of cell death. Remedies capable of averting iron-dependent lipid peroxidation, therefore, are lipophilic antioxidants, including vitamin E, ferrostatin-1 (Fer-1), liproxstatin-1 (Lip-1) and possibly potent bioactive polyphenols. -

Single-Cell RNA-Seq Analysis of the Brainstem of Mutant SOD1 Mice Reveals Perturbed Cell Types and Pathways of Amyotrophic Lateral Sclerosis
HHS Public Access Author manuscript Author ManuscriptAuthor Manuscript Author Neurobiol Manuscript Author Dis. Author manuscript; Manuscript Author available in PMC 2020 September 26. Published in final edited form as: Neurobiol Dis. 2020 July ; 141: 104877. doi:10.1016/j.nbd.2020.104877. Single-cell RNA-seq analysis of the brainstem of mutant SOD1 mice reveals perturbed cell types and pathways of amyotrophic lateral sclerosis Wenting Liua, Sharmila Venugopala, Sana Majida, In Sook Ahna, Graciel Diamantea, Jason Honga, Xia Yanga,b,c,*, Scott H. Chandlera,b,* aDepartment of Integrative Biology & Physiology, University of California, 2024 Terasaki Bld, 610 Charles E. Young Dr. East, Los Angeles, USA bBrain Research Institute, University of California, Los Angeles, USA cInstitute for Quantitative and Computational Biosciences, University of California, Los Angeles, USA Abstract Amyotrophic lateral sclerosis (ALS) is a neurodegenerative disease in which motor neurons throughout the brain and spinal cord progressively degenerate resulting in muscle atrophy, paralysis and death. Recent studies using animal models of ALS implicate multiple cell-types (e.g., astrocytes and microglia) in ALS pathogenesis in the spinal motor systems. To ascertain cellular vulnerability and cell-type specific mechanisms of ALS in the brainstem that orchestrates oral-motor functions, we conducted parallel single cell RNA sequencing (scRNA-seq) analysis using the high-throughput Drop-seq method. We isolated 1894 and 3199 cells from the brainstem of wildtype and mutant SOD1 symptomatic mice respectively, at postnatal day 100. We recovered major known cell types and neuronal subpopulations, such as interneurons and motor neurons, and trigeminal ganglion (TG) peripheral sensory neurons, as well as, previously uncharacterized interneuron subtypes. -

Diallyl Disulfide Inhibits the Proliferation of HT-29 Human Colon Cancer Cells by Inducing Differentially Expressed Genes
MOLECULAR MEDICINE REPORTS 4: 553-559, 2011 Diallyl disulfide inhibits the proliferation of HT-29 human colon cancer cells by inducing differentially expressed genes YOU-SHENG HUANG1,2, NA XIE1,2, QI SU1, JIAN SU1, CHEN HUANG1 and QIAN-JIN LIAO1 1Cancer Research Institute, University of South China, Hengyang, Hunan 421001; 2Department of Pathology, Hainan Medical University, Haikou, Hainan 571101, P.R. China Received November 22, 2010; Accepted February 28, 2011 DOI: 10.3892/mmr.2011.453 Abstract. Diallyl disulfide (DADS), a sulfur compound Introduction derived from garlic, has been shown to have protective effects against colon carcinogenesis in several studies performed in Colon cancer is one of the major causes of cancer death rodent models. However, its molecular mechanism of action worldwide (1). An understanding of the mechanisms involved remains unclear. This study was designed to confirm the anti- in the occurrence and development of colon cancer would aid proliferative activity of DADS and to screen for differentially in its therapy. Epidemiological investigations have provided expressed genes induced by DADS in human colon cancer cells compelling evidence that environmental factors are modifiers with the aim of exploring its possible anticancer mechanisms. in colon cancer (1-3); diet has also been shown to be an impor- The anti-proliferative capability of DADS in the HT-29 human tant determinant of cancer risk and tumor behavior (3-5). colon cancer cells was analyzed by MTT assays and flow Garlic consumption is very popular all over the world. cytometry. The differences in gene expression between DADS- Epidemiological studies have shown an inverse correlation treated (experimental group) and untreated (control group) between the consumption of garlic and colon cancer in certain HT-29 cells were identified using two-directional suppression areas (6). -

Differential Expression of Thymic DNA Repair Genes in Low-Dose-Rate Irradiated AKR/J Mice
pISSN 1229-845X, eISSN 1976-555X JOURNAL OF J. Vet. Sci. (2013), 14(3), 271-279 http://dx.doi.org/10.4142/jvs.2013.14.3.271 Veterinary Received: 9 Mar. 2012, Revised: 19 Sep. 2012, Accepted: 23 Oct. 2012 Science Original Article Differential expression of thymic DNA repair genes in low-dose-rate irradiated AKR/J mice Jin Jong Bong1, Yu Mi Kang1, Suk Chul Shin1, Seung Jin Choi1, Kyung Mi Lee2,*, Hee Sun Kim1,* 1Radiation Health Research Institute, Korea Hydro and Nuclear Power, Seoul 132-703, Korea 2Global Research Lab, BAERI Institute, Department of Biochemistry and Molecular Biology, Korea University College of Medicine, Seoul 136-705, Korea We previously determined that AKR/J mice housed in a harmful to human life. For example, LDR irradiation low-dose-rate (LDR) (137Cs, 0.7 mGy/h, 2.1 Gy) γ-irradiation therapy for cancer is known to stimulate antioxidant facility developed less spontaneous thymic lymphoma and capacity, repair of DNA damage, apoptosis, and induction survived longer than those receiving sham or high-dose-rate of immune responses [4], and LDR irradiation has been (HDR) (137Cs, 0.8 Gy/min, 4.5 Gy) radiation. Interestingly, shown to mitigate lymphomagenesis and prolong the life histopathological analysis showed a mild lymphomagenesis in span of AKR/J mice [17,18]. Conversely, low-dose (LD) the thymus of LDR-irradiated mice. Therefore, in this study, we irradiation is reportedly involved in carcinogenesis, and LD investigated whether LDR irradiation could trigger the irradiation (lower than 2.0 Gy) has been shown to promote expression of thymic genes involved in the DNA repair process tumor growth and metastasis by enhancing angiogenesis of AKR/J mice. -

A Strategy to Address Dissociation-Induced Compositional and Transcriptional Bias for Single-Cell Analysis of the Human Mammary Gland
bioRxiv preprint doi: https://doi.org/10.1101/2021.02.11.430721; this version posted February 11, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. A strategy to address dissociation-induced compositional and transcriptional bias for single-cell analysis of the human mammary gland Lisa K. Engelbrecht1,9,*,‡, Alecia-Jane Twigger1,2,*, Hilary M. Ganz1, Christian J. Gabka4, Andreas R. Bausch5,8, Heiko Lickert6,7,8, Michael Sterr6, Ines Kunze6, Walid T. Khaled2,3 & Christina H. Scheel1,10,‡ Affiliations 1 Institute of Stem Cell Research, Helmholtz Center for Health and Environmental Research Munich, Neuherberg 85764, Germany 2 Department of Pharmacology, University of Cambridge, Cambridge CB2 1PD, UK 3 Cancer Research UK Cambridge Cancer Center, Cambridge CB2 0RE, UK 4 Nymphenburg Clinic for Plastic and Aesthetic Surgery, Munich 80637, Germany 5 Chair of Biophysics E27, Technical University Munich, 85747 Garching, Germany 6 Institute of Diabetes and Regeneration Research, Helmholtz Center for Health and Environmental Research Munich, Neuherberg 85764, Germany 7 German Center for Diabetes Research (DZD), 85764 Neuherberg, Germany 8 Technical University of Munich, 80333 Munich, Germany. 9 Present address: Chair of Biophysics E27, Technical University Munich, 85747 Garching, Germany 10 Present address: Department of Dermatology, St. Josef Hospital, Ruhr-University Bochum, 44791 Bochum, Germany * These authors contributed equally to this work. ‡ Corresponding author. Correspondence to: [email protected] or [email protected] 1 bioRxiv preprint doi: https://doi.org/10.1101/2021.02.11.430721; this version posted February 11, 2021. -

Iron Control of Erythroid Microtubule Cytoskeleton As a Potential Target in Treatment of Iron-Restricted Anemia ✉ Adam N
ARTICLE https://doi.org/10.1038/s41467-021-21938-2 OPEN Iron control of erythroid microtubule cytoskeleton as a potential target in treatment of iron-restricted anemia ✉ Adam N. Goldfarb 1 , Katie C. Freeman1, Ranjit K. Sahu1, Kamaleldin E. Elagib1, Maja Holy1, Abhinav Arneja1, Renata Polanowska-Grabowska2, Alejandro A. Gru 1, Zollie White III1, Shadi Khalil3, Michael J. Kerins4, Aikseng Ooi4, Norbert Leitinger2, Chance John Luckey1 & Lorrie L. Delehanty1 1234567890():,; Anemias of chronic disease and inflammation (ACDI) result from restricted iron delivery to erythroid progenitors. The current studies reveal an organellar response in erythroid iron restriction consisting of disassembly of the microtubule cytoskeleton and associated Golgi disruption. Isocitrate supplementation, known to abrogate the erythroid iron restriction response, induces reassembly of microtubules and Golgi in iron deprived progenitors. Ferritin, based on proteomic profiles, regulation by iron and isocitrate, and putative interaction with microtubules, is assessed as a candidate mediator. Knockdown of ferritin heavy chain (FTH1) in iron replete progenitors induces microtubule collapse and erythropoietic blockade; con- versely, enforced ferritin expression rescues erythroid differentiation under conditions of iron restriction. Fumarate, a known ferritin inducer, synergizes with isocitrate in reversing mole- cular and cellular defects of iron restriction and in oral remediation of murine anemia. These findings identify a cytoskeletal component of erythroid iron restriction and demonstrate potential for its therapeutic targeting in ACDI. 1 Department of Pathology, University of Virginia School of Medicine, Charlottesville, VA, USA. 2 Department of Pharmacology, University of Virginia School of Medicine, Charlottesville, VA, USA. 3 Department of Dermatology, University of California, San Diego School of Medicine, San Diego, CA, USA. -

Discovery of Industrially Relevant Oxidoreductases
DISCOVERY OF INDUSTRIALLY RELEVANT OXIDOREDUCTASES Thesis Submitted for the Degree of Master of Science by Kezia Rajan, B.Sc. Supervised by Dr. Ciaran Fagan School of Biotechnology Dublin City University Ireland Dr. Andrew Dowd MBio Monaghan Ireland January 2020 Declaration I hereby certify that this material, which I now submit for assessment on the programme of study leading to the award of Master of Science, is entirely my own work, and that I have exercised reasonable care to ensure that the work is original, and does not to the best of my knowledge breach any law of copyright, and has not been taken from the work of others save and to the extent that such work has been cited and acknowledged within the text of my work. Signed: ID No.: 17212904 Kezia Rajan Date: 03rd January 2020 Acknowledgements I would like to thank the following: God, for sending me angels in the form of wonderful human beings over the last two years to help me with any- and everything related to my project. Dr. Ciaran Fagan and Dr. Andrew Dowd, for guiding me and always going out of their way to help me. Thank you for your patience, your advice, and thank you for constantly believing in me. I feel extremely privileged to have gotten an opportunity to work alongside both of you. Everything I’ve learnt and the passion for research that this project has sparked in me, I owe it all to you both. Although I know that words will never be enough to express my gratitude, I still want to say a huge thank you from the bottom of my heart.