Pa28αβ: the Enigmatic Magic Ring of the Proteasome?
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Antigen Presentation by Dendritic Cells and Their Significance in Antineoplastic Immunotherapy
in vivo 18: 81-100 (2004) Antigen Presentation by Dendritic Cells and their Significance in Antineoplastic Immunotherapy BELA BODEY1,2, STUART E. SIEGEL1,2 and HANS E. KAISER3 1Department of Pathology, Keck School of Medicine, University of Southern California, Los Angeles, CA; 2Childrens’ Center for Cancer and Blood Diseases, Childrens’ Hospital Los Angeles, Los Angeles, CA; 3Department of Pathology, School of Medicine, University of Maryland, Baltimore, MD, U.S.A. and Department of General and Experimental Pathology, University of Vienna, Vienna, Austria Abstract. Dendritic cells (DCs) are present in essentially every derived peptides on the surface of major histocompatibility mammalian tissue, where they operate at the interface of innate complex (MHC) molecules and acquire the cellular specialization and acquired immunity by recognizing pathogens and presenting to select and activate naive antigen-specific T lymphocytes. pathogen-derived peptides to T lymphocytes. According to the Immunotherapeutic ideas are based on the ability of the research group of Shortman (1-9), experimental results suggest a mammalian immune system to recognize neoplastically "dual" DC differentiation model, demonstrating the existence of transformed cells. Immunotherapy of human neoplasms has both myeloid-derived (with characteristic IF: CD11b+, CD11c+, always represented a very attractive fourth-modality therapeutic CD8alpha- and DEC205+) and lymphoid-derived DCs (showing approach, especially in light of the many shortcomings of CD11b-, CD11c-, CD8alpha+ and DEC205+ IF). DCs, including conventional surgical, radiation and chemotherapies in the interdigitating cells (IDCs) and Langerhans cells (LCs), are management of neoplastically transformed cells. The cancer characterized by dendritic morphology, high migratory mobility and vaccine approach to therapy is based on the notion that the are the most effective, "professional" cells for antigen presentation immune system could possibly mount a rejection strength response in primary immune responses. -

The Global Architecture Shaping the Heterogeneity and Tissue-Dependency of the MHC Class I Immunopeptidome Is Evolutionarily Conserved
bioRxiv preprint doi: https://doi.org/10.1101/2020.09.28.317750; this version posted September 29, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. The Global Architecture Shaping the Heterogeneity and Tissue-Dependency of the MHC Class I Immunopeptidome is Evolutionarily Conserved Authors Peter Kubiniok†1, Ana Marcu†2,3, Leon Bichmann†2,4, Leon Kuchenbecker4, Heiko Schuster1,5, David Hamelin1, Jérome Despault1, Kevin Kovalchik1, Laura Wessling1, Oliver Kohlbacher4,7,8,9,10 Stefan Stevanovic2,3,6, Hans-Georg Rammensee2,3,6, Marian C. Neidert11, Isabelle Sirois1, Etienne Caron1,12* Affiliations *Corresponding and Leading author: Etienne Caron ([email protected]) †Equal contribution to this work 1CHU Sainte-Justine Research Center, Montreal, QC H3T 1C5, Canada 2Department of Immunology, Interfaculty Institute for Cell Biology, University of Tübingen, Tübingen, Baden-Württemberg, 72076, Germany. 3Cluster of Excellence iFIT (EXC 2180) "Image-Guided and Functionally Instructed Tumor Therapies", University of Tübingen, Tübingen, Baden-Württemberg, 72076, Germany. 4Applied Bioinformatics, Dept. of Computer Science, University of Tübingen, Tübingen, Baden- Württemberg, 72074, Germany. 5Immatics Biotechnologies GmbH, Tübingen, 72076, Baden-Württemberg, Germany. 6DKFZ Partner Site Tübingen, German Cancer Consortium (DKTK), Tübingen, Baden- Württemberg, 72076, Germany. 7Institute for Bioinformatics and Medical Informatics, -

Antigen Processing (Major Hbitocompatlbility Complex/Class I Moecules/Lymphokines) YOUNG YANG, JAMES B
Proc. Nati. Acad. Sci. USA Vol. 89, pp. 4928-4932, June 1992 Immunology Proteasomes are regulated by interferon y: Implications for antigen processing (major hbitocompatlbility complex/class I moecules/lymphokines) YOUNG YANG, JAMES B. WATERS, KLAUS FROH, AND PER A. PETERSON Department of Immunology, The Scripps Research Institute, La Jolla, CA 92037 Communicated by Frank J. Dixon, February 27, 1992 ABSTRACT Class I major histocompatibility complex strengthened by the findings, presented in this communica- (MHC) molecules present antigenic peptides of cytoplasmic tion, that several proteasomal subunits, including MHC- origin to T cells. As the lengths ofthese peptides seem stried encoded subunits, are regulated by interferon y (IFN--y) and to eight or nine amino acids, an unusual proteolytic system that the incorporation of several more subunits into protea- must play a role in antigen processing. Proteasomes, a major somes appears to depend on the expression of the MHC- extralysosomal proteolytic system, are responsible for the encoded proteasomal subunits. Moreover, the pattern of degradation of cytoplasmic proteins. We demonstrate that expression of IFN-y-regulated subunits suggests complexi- several proteasomal subunits, including MHC-encoded sub- ties in the regulation of proteasomes with respect to its units, are regulated by interferon y. These data and the finding subunit composition, subcellular localization, and its incor- that MHC-encoded and other interferon -regulated protea- poration into larger ubiquitin-related proteolytic complexes. somal subunits are uniquely associated with proteasomes Possible functions for the MHC-encoded and IFN-y- strongly suggest that the immune system has recruited protea- regulated proteasomal subunits in antigen processing are somes for antigen processing. -

Cytoplasmic Proteolysis Antigens on MHC Class II Is Dependent On
Efficient Presentation of Both Cytosolic and Endogenous Transmembrane Protein Antigens on MHC Class II Is Dependent on Cytoplasmic Proteolysis This information is current as of September 28, 2021. Paushali Mukherjee, Aadish Dani, Sumeena Bhatia, Nagendra Singh, Alexander Y. Rudensky, Anna George, Vineeta Bal, Satyajit Mayor and Satyajit Rath J Immunol 2001; 167:2632-2641; ; doi: 10.4049/jimmunol.167.5.2632 Downloaded from http://www.jimmunol.org/content/167/5/2632 References This article cites 51 articles, 20 of which you can access for free at: http://www.jimmunol.org/content/167/5/2632.full#ref-list-1 http://www.jimmunol.org/ Why The JI? Submit online. • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists by guest on September 28, 2021 • Fast Publication! 4 weeks from acceptance to publication *average Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2001 by The American Association of Immunologists All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. Efficient Presentation of Both Cytosolic and Endogenous Transmembrane Protein Antigens on MHC Class II Is Dependent on Cytoplasmic Proteolysis1 Paushali Mukherjee,* Aadish Dani,† Sumeena Bhatia,* Nagendra Singh,* Alexander Y. -

On the Role of the Immunoproteasome in Transplant Rejection
Immunogenetics (2019) 71:263–271 https://doi.org/10.1007/s00251-018-1084-0 REVIEW On the role of the immunoproteasome in transplant rejection Michael Basler1,2 & Jun Li1,3 & Marcus Groettrup1,2 Received: 17 July 2018 /Accepted: 4 September 2018 /Published online: 15 September 2018 # Springer-Verlag GmbH Germany, part of Springer Nature 2018 Abstract The immunoproteasome is expressed in cells of hematopoietic origin and is induced during inflammation by IFN-γ. Targeting the immunoproteasome with selective inhibitors has been shown to be therapeutically effective in pre-clinical models for autoim- mune diseases, colitis-associated cancer formation, and transplantation. Immunoproteasome inhibition prevents activation and proliferation of lymphocytes, lowers MHC class I cell surface expression, reduces the expression of cytokines of activated immune cells, and curtails T helper 1 and 17 cell differentiation. This might explain the in vivo efficacy of immunoproteasome inhibition in different pre-clinical disease models for autoimmunity, cancer, and transplantation. In this review, we summarize the effect of immunoproteasome inhibition in different animal models for transplantation. Keywords Proteasome . Immunoproteasome . Antigen processing . Antigen presentation . Transplantation Introduction et al. 2012). Depending on the cell type and the presence or absence of the pro-inflammatory cytokine interferon (IFN)-γ, The proteasome is responsible for the degradation of proteins the three inducible β subunits of the immunoproteasome low in the cytoplasm and nuclei of all eukaryotic cells and exerts molecular mass polypeptide (LMP)2 (β1i), multicatalytic en- numerous essential regulatory functions in nearly all cell bio- dopeptidase complex-like (MECL)-1 (β2i), and LMP7 (β5i), logical pathways. The 26S proteasome degrades poly- can, in addition to the corresponding constitutive subunits ubiquitylated protein substrates and consists of a 19S regulator β1c, β2c, and β5c, enrich the cellular assortment of catalyti- and a 20S proteolytic core complex. -
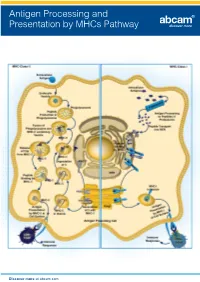
Antigen Processing and Presentation by Mhcs Pathway
Antigen Processing and Presentation by MHCs Pathway Discover more at abcam.com Antigen Processing and Presentation by MHCs Pathway Related antibodies Product This image shows MHC Class II highlights Product Clonality Applications Host Species Reactivity Product code expressing cells in formalin fixed and paraffin embedded human lymph MHC class I antibody [EP1395Y] M Flow Cyt, ICC/IF, IHC-P, IP, WB Rb Hu, Ms, Rat 52922 node, using ab55152. MHC class I antibody [AF6-88.5.5.3] (Biotin) M Flow Cyt Ms Ms 93528 MHC Class II antibody MHC class I antibody [34-1-2S] (FITC) M Flow Cyt Ms Ms 95572 (ab55152) MHC class I antibody [34-1-2S] (Phycoerythrin) M Flow Cyt Ms Ms 95571 MHC Class I alpha antibody [F21-2] M AP, Flow Cyt, IP, WB Ms Chk 23483 Clonality Applications MHC Class 1 H2 Db antibody [27-11-13] - BSA and Azide free M Flow Cyt, FuncS, IHC-Fr Ms Ms 25244 M IHC-P, WB MHC Class 1 H2 Db antibody [28-14-8] (FITC) M Flow Cyt Ms Ms 25056 Host Species cross reactivity MHC Class 1 H2 Db antibody [KH95] M Flow Cyt, FuncS, WB Ms Ms 64373 Ms Hu, RecFrag MHC Class I H2 Dd antibody [34-2-12] M Flow Cyt, FuncS, IHC-Fr, IP Ms Ms 64368 MHC Class I H2 Dk antibody [15-5-5.3] M Flow Cyt Ms Ms 25216 MHC Class II antigens are a valuable tool for studying T helper cell interactions with class II MHC Class I H2 Kb antibody P ELISA, WB Rb Ms 93364 positive antigen presenting cells, as well as the MHC Class I H2 Kd + Dd + q + u + v antibody [1.B.552] (FITC) M Flow Cyt, IP Ms Ms 62228 development of T helper cells since the antigen is present on stromal cells in the thymus. -

Supplemental Materials Supplemental Table 1
Electronic Supplementary Material (ESI) for RSC Advances. This journal is © The Royal Society of Chemistry 2016 Supplemental Materials Supplemental Table 1. The differentially expressed proteins from rat pancreas identified by proteomics (SAP vs. SO) No. Protein name Gene name ratio P value 1 Metallothionein Mt1m 3.35 6.34E-07 2 Neutrophil antibiotic peptide NP-2 Defa 3.3 8.39E-07 3 Ilf2 protein Ilf2 3.18 1.75E-06 4 Numb isoform o/o rCG 3.12 2.73E-06 5 Lysozyme Lyz2 3.01 5.63E-06 6 Glucagon Gcg 2.89 1.17E-05 7 Serine protease HTRA1 Htra1 2.75 2.97E-05 8 Alpha 2 macroglobulin cardiac isoform (Fragment) 2.75 2.97E-05 9 Myosin IF (Predicted) Myo1f 2.65 5.53E-05 10 Neuroendocrine secretory protein 55 Gnas 2.61 7.60E-05 11 Matrix metallopeptidase 8 Mmp8 2.57 9.47E-05 12 Protein Tnks1bp1 Tnks1bp1 2.53 1.22E-04 13 Alpha-parvin Parva 2.47 1.78E-04 14 C4b-binding protein alpha chain C4bpa 2.42 2.53E-04 15 Protein KTI12 homolog Kti12 2.41 2.74E-04 16 Protein Rab11fip5 Rab11fip5 2.41 2.84E-04 17 Protein Mcpt1l3 Mcpt1l3 2.33 4.43E-04 18 Phospholipase B-like 1 Plbd1 2.33 4.76E-04 Aldehyde dehydrogenase (NAD), cytosolic 19 2.32 4.93E-04 (Fragments) 20 Protein Dpy19l2 Dpy19l2 2.3 5.68E-04 21 Regenerating islet-derived 3 alpha, isoform CRA_a Reg3a 2.27 6.74E-04 22 60S acidic ribosomal protein P1 Rplp1 2.26 7.22E-04 23 Serum albumin Alb 2.25 7.98E-04 24 Ribonuclease 4 Rnase4 2.24 8.25E-04 25 Cct-5 protein (Fragment) Cct5 2.24 8.52E-04 26 Protein S100-A9 S100a9 2.22 9.71E-04 27 Creatine kinase M-type Ckm 2.21 1.00E-03 28 Protein Larp4b Larp4b 2.18 1.25E-03 -

Leishmania (L.) Amazonensis Peptidase Activities Inside the Living Cells and in Their Lysates
Molecular & Biochemical Parasitology 184 (2012) 82–89 Contents lists available at SciVerse ScienceDirect Molecular & Biochemical Parasitology Leishmania (L.) amazonensis peptidase activities inside the living cells and in their lysates a a b c a Elide E. Caroselli , Diego M. Assis , Clara L. Barbiéri , Wagner A.S. Júdice , Maria A. Juliano , d a,∗ Marcos L. Gazarini , Luiz Juliano a Department of Biophysics, Escola Paulista de Medicina, Universidade Federal de São Paulo, SP, Brazil b Department of Microbiology, Immunology and Parasitology, Escola Paulista de Medicina, Universidade Federal de São Paulo, SP, Brazil c Centro Interdisciplinar de Investigac¸ ão Bioquímica, Universidade de Mogi das Cruzes, Av. Dr. Cândido Xavier de Almeida Souza 200, 08780-911 Mogi das Cruzes, Brazil d Department of Biosciences, Universidade Federal de São Paulo, Santos, Brazil a r t i c l e i n f o a b s t r a c t Article history: In this study we investigated the peptidase activity in Leishmania (L.) amazonensis live amastigote by con- Received 24 November 2011 focal microscopy using peptidyl-MCA as substrates, the hydrolysis of which releases the MCA fluorophore Received in revised form 13 March 2012 inside the cells. Cell pre-treatment with peptidase inhibitors indicated the presence of cysteine and ser- Accepted 27 April 2012 ine peptidases. It was noteworthy that Leishmania amastigotes incorporate only substrates (Z-FR-MCA, Available online 6 May 2012 Z-RR-MCA) or inhibitors (E64, TLCK) containing positively charged groups. The peptidase activities in the supernatants of amastigotes and promastigotes lysates were also evaluated with the same peptidyl-MCA Keywords: substrates and inhibitors in the pH range 4.5–9.0. -

Supplementary Table 9. Functional Annotation Clustering Results for the Union (GS3) of the Top Genes from the SNP-Level and Gene-Based Analyses (See ST4)
Supplementary Table 9. Functional Annotation Clustering Results for the union (GS3) of the top genes from the SNP-level and Gene-based analyses (see ST4) Column Header Key Annotation Cluster Name of cluster, sorted by descending Enrichment score Enrichment Score EASE enrichment score for functional annotation cluster Category Pathway Database Term Pathway name/Identifier Count Number of genes in the submitted list in the specified term % Percentage of identified genes in the submitted list associated with the specified term PValue Significance level associated with the EASE enrichment score for the term Genes List of genes present in the term List Total Number of genes from the submitted list present in the category Pop Hits Number of genes involved in the specified term (category-specific) Pop Total Number of genes in the human genome background (category-specific) Fold Enrichment Ratio of the proportion of count to list total and population hits to population total Bonferroni Bonferroni adjustment of p-value Benjamini Benjamini adjustment of p-value FDR False Discovery Rate of p-value (percent form) Annotation Cluster 1 Enrichment Score: 3.8978262119731335 Category Term Count % PValue Genes List Total Pop Hits Pop Total Fold Enrichment Bonferroni Benjamini FDR GOTERM_CC_DIRECT GO:0005886~plasma membrane 383 24.33290978 5.74E-05 SLC9A9, XRCC5, HRAS, CHMP3, ATP1B2, EFNA1, OSMR, SLC9A3, EFNA3, UTRN, SYT6, ZNRF2, APP, AT1425 4121 18224 1.18857065 0.038655922 0.038655922 0.086284383 UP_KEYWORDS Membrane 626 39.77128335 1.53E-04 SLC9A9, HRAS, -

The Role of the Immunoproteasome in Interferon-Γ-Mediated Microglial Activation Received: 4 May 2017 Kasey E
www.nature.com/scientificreports OPEN The role of the immunoproteasome in interferon-γ-mediated microglial activation Received: 4 May 2017 Kasey E. Moritz1, Nikki M. McCormack1, Mahlet B. Abera2, Coralie Viollet3, Young J. Yauger1, Accepted: 14 July 2017 Gauthaman Sukumar3, Clifton L. Dalgard1,2,3,4 & Barrington G. Burnett 1,2 Published: xx xx xxxx Microglia regulate the brain microenvironment by sensing damage and neutralizing potentially harmful insults. Disruption of central nervous system (CNS) homeostasis results in transition of microglia to a reactive state characterized by morphological changes and production of cytokines to prevent further damage to CNS tissue. Immunoproteasome levels are elevated in activated microglia in models of stroke, infection and traumatic brain injury, though the exact role of the immunoproteasome in neuropathology remains poorly defned. Using gene expression analysis and native gel electrophoresis we characterize the expression and assembly of the immunoproteasome in microglia following interferon-gamma exposure. Transcriptome analysis suggests that the immunoproteasome regulates multiple features of microglial activation including nitric oxide production and phagocytosis. We show that inhibiting the immunoproteasome attenuates expression of pro-infammatory cytokines and suppresses interferon-gamma-dependent priming of microglia. These results imply that targeting immunoproteasome function following CNS injury may attenuate select microglial activity to improve the pathophysiology of neurodegenerative conditions or the progress of infammation-mediated secondary injury following neurotrauma. Microglia are the primary infammatory mediators of the central nervous system (CNS). Damage to the CNS results in the transition of microglia from a surveying or ‘ramifed’ state, to a ‘reactive’ state, allowing them to respond to changes in the local milieu1–3. -

Mechanisms of HIV Protein Degradation Into Epitopes: Implications for Vaccine Design
Viruses 2014, 6, 3271-3292; doi:10.3390/v6083271 OPEN ACCESS viruses ISSN 1999-4915 www.mdpi.com/journal/viruses Review Mechanisms of HIV Protein Degradation into Epitopes: Implications for Vaccine Design Marijana Rucevic †, Julie Boucau †, Jens Dinter †, Georgio Kourjian † and Sylvie Le Gall * Ragon Institute of MGH, MIT and Harvard, Massachusetts General Hospital and Harvard Medical School, Cambridge, MA 02139, USA; E-Mails: [email protected] (M.R.); [email protected] (J.B.); [email protected] (J.D.); [email protected] (G.K.) † These authors contributed equally to this work. * Author to whom correspondence should be addressed; E-Mail: [email protected]; Tel.: +1-857-268-7010. Received: 11 June 2014; in revised form: 6 August 2014 / Accepted: 11 August 2014 / Published: 21 August 2014 Abstract: The degradation of HIV-derived proteins into epitopes displayed by MHC-I or MHC-II are the first events leading to the priming of HIV-specific immune responses and to the recognition of infected cells. Despite a wealth of information about peptidases involved in protein degradation, our knowledge of epitope presentation during HIV infection remains limited. Here we review current data on HIV protein degradation linking epitope production and immunodominance, viral evolution and impaired epitope presentation. We propose that an in-depth understanding of HIV antigen processing and presentation in relevant primary cells could be exploited to identify signatures leading to efficient or inefficient epitope presentation in HIV proteomes, and to improve the design of immunogens eliciting immune responses efficiently recognizing all infected cells. Keywords: HIV; antigen processing; protein degradation; proteasome; aminopeptidase; peptidase; immunogen; vaccine vector; dendritic cells; T cells; viral evolution Viruses 2014, 6 3272 1. -

The Microbiota-Produced N-Formyl Peptide Fmlf Promotes Obesity-Induced Glucose
Page 1 of 230 Diabetes Title: The microbiota-produced N-formyl peptide fMLF promotes obesity-induced glucose intolerance Joshua Wollam1, Matthew Riopel1, Yong-Jiang Xu1,2, Andrew M. F. Johnson1, Jachelle M. Ofrecio1, Wei Ying1, Dalila El Ouarrat1, Luisa S. Chan3, Andrew W. Han3, Nadir A. Mahmood3, Caitlin N. Ryan3, Yun Sok Lee1, Jeramie D. Watrous1,2, Mahendra D. Chordia4, Dongfeng Pan4, Mohit Jain1,2, Jerrold M. Olefsky1 * Affiliations: 1 Division of Endocrinology & Metabolism, Department of Medicine, University of California, San Diego, La Jolla, California, USA. 2 Department of Pharmacology, University of California, San Diego, La Jolla, California, USA. 3 Second Genome, Inc., South San Francisco, California, USA. 4 Department of Radiology and Medical Imaging, University of Virginia, Charlottesville, VA, USA. * Correspondence to: 858-534-2230, [email protected] Word Count: 4749 Figures: 6 Supplemental Figures: 11 Supplemental Tables: 5 1 Diabetes Publish Ahead of Print, published online April 22, 2019 Diabetes Page 2 of 230 ABSTRACT The composition of the gastrointestinal (GI) microbiota and associated metabolites changes dramatically with diet and the development of obesity. Although many correlations have been described, specific mechanistic links between these changes and glucose homeostasis remain to be defined. Here we show that blood and intestinal levels of the microbiota-produced N-formyl peptide, formyl-methionyl-leucyl-phenylalanine (fMLF), are elevated in high fat diet (HFD)- induced obese mice. Genetic or pharmacological inhibition of the N-formyl peptide receptor Fpr1 leads to increased insulin levels and improved glucose tolerance, dependent upon glucagon- like peptide-1 (GLP-1). Obese Fpr1-knockout (Fpr1-KO) mice also display an altered microbiome, exemplifying the dynamic relationship between host metabolism and microbiota.