Facial Dysostosis Information Sheet 6-10-19
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Syndromic Ear Anomalies and Renal Ultrasounds
Syndromic Ear Anomalies and Renal Ultrasounds Raymond Y. Wang, MD*; Dawn L. Earl, RN, CPNP‡; Robert O. Ruder, MD§; and John M. Graham, Jr, MD, ScD‡ ABSTRACT. Objective. Although many pediatricians cific MCA syndromes that have high incidences of renal pursue renal ultrasonography when patients are noted to anomalies. These include CHARGE association, Townes- have external ear malformations, there is much confusion Brocks syndrome, branchio-oto-renal syndrome, Nager over which specific ear malformations do and do not syndrome, Miller syndrome, and diabetic embryopathy. require imaging. The objective of this study was to de- Patients with auricular anomalies should be assessed lineate characteristics of a child with external ear malfor- carefully for accompanying dysmorphic features, includ- mations that suggest a greater risk of renal anomalies. We ing facial asymmetry; colobomas of the lid, iris, and highlight several multiple congenital anomaly (MCA) retina; choanal atresia; jaw hypoplasia; branchial cysts or syndromes that should be considered in a patient who sinuses; cardiac murmurs; distal limb anomalies; and has both ear and renal anomalies. imperforate or anteriorly placed anus. If any of these Methods. Charts of patients who had ear anomalies features are present, then a renal ultrasound is useful not and were seen for clinical genetics evaluations between only in discovering renal anomalies but also in the diag- 1981 and 2000 at Cedars-Sinai Medical Center in Los nosis and management of MCA syndromes themselves. Angeles and Dartmouth-Hitchcock Medical Center in A renal ultrasound should be performed in patients with New Hampshire were reviewed retrospectively. Only pa- isolated preauricular pits, cup ears, or any other ear tients who underwent renal ultrasound were included in anomaly accompanied by 1 or more of the following: the chart review. -
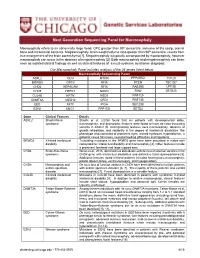
Macrocephaly Information Sheet 6-13-19
Next Generation Sequencing Panel for Macrocephaly Clinical Features: Macrocephaly refers to an abnormally large head, OFC greater than 98th percentile, inclusive of the scalp, cranial bone and intracranial contents. Megalencephaly, brain weight/volume ratio greater than 98th percentile, results from true enlargement of the brain parenchyma [1]. Megalencephaly is typically accompanied by macrocephaly, however macrocephaly can occur in the absence of megalencephaly [2]. Both macrocephaly and megalencephaly can been seen as isolated clinical findings as well as clinical features of a mutli-systemic syndromic diagnosis. Our Macrocephaly Panel includes analysis of the 36 genes listed below. Macrocephaly Sequencing Panel ASXL2 GLI3 MTOR PPP2R5D TCF20 BRWD3 GPC3 NFIA PTEN TBC1D7 CHD4 HEPACAM NFIX RAB39B UPF3B CHD8 HERC1 NONO RIN2 ZBTB20 CUL4B KPTN NSD1 RNF125 DNMT3A MED12 OFD1 RNF135 EED MITF PIGA SEC23B EZH2 MLC1 PPP1CB SETD2 Gene Clinical Features Details ASXL2 Shashi-Pena Shashi et al. (2016) found that six patients with developmental delay, syndrome macrocephaly, and dysmorphic features were found to have de novo truncating variants in ASXL2 [3]. Distinguishing features were macrocephaly, absence of growth retardation, and variability in the degree of intellectual disabilities The phenotype also consisted of prominent eyes, arched eyebrows, hypertelorism, a glabellar nevus flammeus, neonatal feeding difficulties and hypotonia. BRWD3 X-linked intellectual Truncating mutations in the BRWD3 gene have been described in males with disability nonsyndromic intellectual disability and macrocephaly [4]. Other features include a prominent forehead and large cupped ears. CHD4 Sifrim-Hitz-Weiss Weiss et al., 2016, identified five individuals with de novo missense variants in the syndrome CHD4 gene with intellectual disabilities and distinctive facial dysmorphisms [5]. -

Blueprint Genetics Craniosynostosis Panel
Craniosynostosis Panel Test code: MA2901 Is a 38 gene panel that includes assessment of non-coding variants. Is ideal for patients with craniosynostosis. About Craniosynostosis Craniosynostosis is defined as the premature fusion of one or more cranial sutures leading to secondary distortion of skull shape. It may result from a primary defect of ossification (primary craniosynostosis) or, more commonly, from a failure of brain growth (secondary craniosynostosis). Premature closure of the sutures (fibrous joints) causes the pressure inside of the head to increase and the skull or facial bones to change from a normal, symmetrical appearance resulting in skull deformities with a variable presentation. Craniosynostosis may occur in an isolated setting or as part of a syndrome with a variety of inheritance patterns and reccurrence risks. Craniosynostosis occurs in 1/2,200 live births. Availability 4 weeks Gene Set Description Genes in the Craniosynostosis Panel and their clinical significance Gene Associated phenotypes Inheritance ClinVar HGMD ALPL Odontohypophosphatasia, Hypophosphatasia perinatal lethal, AD/AR 78 291 infantile, juvenile and adult forms ALX3 Frontonasal dysplasia type 1 AR 8 8 ALX4 Frontonasal dysplasia type 2, Parietal foramina AD/AR 15 24 BMP4 Microphthalmia, syndromic, Orofacial cleft AD 8 39 CDC45 Meier-Gorlin syndrome 7 AR 10 19 EDNRB Hirschsprung disease, ABCD syndrome, Waardenburg syndrome AD/AR 12 66 EFNB1 Craniofrontonasal dysplasia XL 28 116 ERF Craniosynostosis 4 AD 17 16 ESCO2 SC phocomelia syndrome, Roberts syndrome -

Polydactyly of the Hand
A Review Paper Polydactyly of the Hand Katherine C. Faust, MD, Tara Kimbrough, BS, Jean Evans Oakes, MD, J. Ollie Edmunds, MD, and Donald C. Faust, MD cleft lip/palate, and spina bifida. Thumb duplication occurs in Abstract 0.08 to 1.4 per 1000 live births and is more common in Ameri- Polydactyly is considered either the most or second can Indians and Asians than in other races.5,10 It occurs in a most (after syndactyly) common congenital hand ab- male-to-female ratio of 2.5 to 1 and is most often unilateral.5 normality. Polydactyly is not simply a duplication; the Postaxial polydactyly is predominant in black infants; it is most anatomy is abnormal with hypoplastic structures, ab- often inherited in an autosomal dominant fashion, if isolated, 1 normally contoured joints, and anomalous tendon and or in an autosomal recessive pattern, if syndromic. A prospec- ligament insertions. There are many ways to classify tive San Diego study of 11,161 newborns found postaxial type polydactyly, and surgical options range from simple B polydactyly in 1 per 531 live births (1 per 143 black infants, excision to complicated bone, ligament, and tendon 1 per 1339 white infants); 76% of cases were bilateral, and 3 realignments. The prevalence of polydactyly makes it 86% had a positive family history. In patients of non-African descent, it is associated with anomalies in other organs. Central important for orthopedic surgeons to understand the duplication is rare and often autosomal dominant.5,10 basic tenets of the abnormality. Genetics and Development As early as 1896, the heritability of polydactyly was noted.11 As olydactyly is the presence of extra digits. -

MR Imaging of Fetal Head and Neck Anomalies
Neuroimag Clin N Am 14 (2004) 273–291 MR imaging of fetal head and neck anomalies Caroline D. Robson, MB, ChBa,b,*, Carol E. Barnewolt, MDa,c aDepartment of Radiology, Children’s Hospital Boston, 300 Longwood Avenue, Harvard Medical School, Boston, MA 02115, USA bMagnetic Resonance Imaging, Advanced Fetal Care Center, Children’s Hospital Boston, Harvard Medical School, 300 Longwood Avenue, Boston, MA 02115, USA cFetal Imaging, Advanced Fetal Care Center, Children’s Hospital Boston, Harvard Medical School, 300 Longwood Avenue, Boston, MA 02115, USA Fetal dysmorphism can occur as a result of var- primarily used for fetal MR imaging. When the fetal ious processes that include malformation (anoma- face is imaged, the sagittal view permits assessment lous formation of tissue), deformation (unusual of the frontal and nasal bones, hard palate, tongue, forces on normal tissue), disruption (breakdown of and mandible. Abnormalities include abnormal promi- normal tissue), and dysplasia (abnormal organiza- nence of the frontal bone (frontal bossing) and lack of tion of tissue). the usual frontal prominence. Abnormal nasal mor- An approach to fetal diagnosis and counseling of phology includes variations in the size and shape of the parents incorporates a detailed assessment of fam- the nose. Macroglossia and micrognathia are also best ily history, maternal health, and serum screening, re- diagnosed on sagittal images. sults of amniotic fluid analysis for karyotype and Coronal images are useful for evaluating the in- other parameters, and thorough imaging of the fetus tegrity of the fetal lips and palate and provide as- with sonography and sometimes fetal MR imaging. sessment of the eyes, nose, and ears. -

Frontonasal Dysplasia
Frontonasal Dysplasia Frontonasal dysplasia is a developmental field defect of cran- f. Ocular anomalies iofacial region characterized by hypertelorism and varying i. Hypertelorism degrees of median nasal clefting. In 1967, DeMeyer first ii. Epicanthal folds described the malformation complex ‘median cleft face syn- iii. Narrowing of the palpebral fissures drome’ to emphasize the key midface defects. Since then several iv. Accessory nasal eyelid tissue with secondary terms have been introduced: frontonasal dysplasia, fron- displacement of inferior puncti colobomas tonasal syndrome, frontonasal dysostosis, and craniofrontonasal v. Epibulbar dermoids dysplasia. vi. Upper eyelid colobomas vii. Microphthalmia GENETICS/BASIC DEFECTS viii. Vitreoretinal degeneration with retinal detachment ix. Congenital cataracts 1. Inheritance g. Facial anomalies a. Sporadic in most cases i. Widow’s peak configuration of the anterior hair- b. Rare autosomal dominant inheritance with variable line in the forehead expression ii. Median cleft of upper lip c. Rare autosomal recessive inheritance iii. Median cleft palate d. Rare X-linked dominant inheritance iv. Preauricular tag 2. Rare association with chromosome anomalies v. Absent tragus a. Partial trisomy 2q and partial monosomy 7q from a vi. Low-set ears balanced maternal t(2;7)(q31;q36) h. Other anomalies b. 22q11 microdeletion i. Conductive deafness c. Reciprocal translocation t(15;22)(q22;q13) ii. Hypoplastic frontal sinuses d. Complex translocation involving chromosomes 3, 7, iii. Cardiac anomalies, especially teratology of and 11 Fallot 3. Rare variants of frontonasal dysplasia/malformation with iv. Limb anomalies variable inheritance patterns a) Clinodactyly 4. Embryologically classified as a developmental field b) Polydactyly defect c) Syndactyly 5. Extreme variable phenotypic expression d) Tibial hypoplasia v. -

Blueprint Genetics Comprehensive Growth Disorders / Skeletal
Comprehensive Growth Disorders / Skeletal Dysplasias and Disorders Panel Test code: MA4301 Is a 374 gene panel that includes assessment of non-coding variants. This panel covers the majority of the genes listed in the Nosology 2015 (PMID: 26394607) and all genes in our Malformation category that cause growth retardation, short stature or skeletal dysplasia and is therefore a powerful diagnostic tool. It is ideal for patients suspected to have a syndromic or an isolated growth disorder or a skeletal dysplasia. About Comprehensive Growth Disorders / Skeletal Dysplasias and Disorders This panel covers a broad spectrum of diseases associated with growth retardation, short stature or skeletal dysplasia. Many of these conditions have overlapping features which can make clinical diagnosis a challenge. Genetic diagnostics is therefore the most efficient way to subtype the diseases and enable individualized treatment and management decisions. Moreover, detection of causative mutations establishes the mode of inheritance in the family which is essential for informed genetic counseling. For additional information regarding the conditions tested on this panel, please refer to the National Organization for Rare Disorders and / or GeneReviews. Availability 4 weeks Gene Set Description Genes in the Comprehensive Growth Disorders / Skeletal Dysplasias and Disorders Panel and their clinical significance Gene Associated phenotypes Inheritance ClinVar HGMD ACAN# Spondyloepimetaphyseal dysplasia, aggrecan type, AD/AR 20 56 Spondyloepiphyseal dysplasia, Kimberley -

Familial Poland Anomaly
J Med Genet: first published as 10.1136/jmg.19.4.293 on 1 August 1982. Downloaded from Journal ofMedical Genetics, 1982, 19, 293-296 Familial Poland anomaly T J DAVID From the Department of Child Health, University of Manchester, Booth Hall Children's Hospital, Manchester SUMMARY The Poland anomaly is usually a non-genetic malformation syndrome. This paper reports two second cousins who both had a typical left sided Poland anomaly, and this constitutes the first recorded case of this condition affecting more than one member of a family. Despite this, for the purposes of genetic counselling, the Poland anomaly can be regarded as a sporadic condition with an extremely low recurrence risk. The Poland anomaly comprises congenital unilateral slightly reduced. The hands were normal. Another absence of part of the pectoralis major muscle in son (Greif himself) said that his own left pectoralis combination with a widely varying spectrum of major was weaker than the right. "Although the ipsilateral upper limb defects.'-4 There are, in difference is obvious, the author still had to carry addition, patients with absence of the pectoralis out his military duties"! major in whom the upper limbs are normal, and Trosev and colleagues9 have been widely quoted as much confusion has been caused by the careless reporting familial cases of the Poland anomaly. labelling of this isolated defect as the Poland However, this is untrue. They described a mother anomaly. It is possible that the two disorders are and child with autosomal dominant radial sided part of a single spectrum, though this has never been upper limb defects. -

Blueprint Genetics Comprehensive Skeletal Dysplasias and Disorders
Comprehensive Skeletal Dysplasias and Disorders Panel Test code: MA3301 Is a 251 gene panel that includes assessment of non-coding variants. Is ideal for patients with a clinical suspicion of disorders involving the skeletal system. About Comprehensive Skeletal Dysplasias and Disorders This panel covers a broad spectrum of skeletal disorders including common and rare skeletal dysplasias (eg. achondroplasia, COL2A1 related dysplasias, diastrophic dysplasia, various types of spondylo-metaphyseal dysplasias), various ciliopathies with skeletal involvement (eg. short rib-polydactylies, asphyxiating thoracic dysplasia dysplasias and Ellis-van Creveld syndrome), various subtypes of osteogenesis imperfecta, campomelic dysplasia, slender bone dysplasias, dysplasias with multiple joint dislocations, chondrodysplasia punctata group of disorders, neonatal osteosclerotic dysplasias, osteopetrosis and related disorders, abnormal mineralization group of disorders (eg hypopohosphatasia), osteolysis group of disorders, disorders with disorganized development of skeletal components, overgrowth syndromes with skeletal involvement, craniosynostosis syndromes, dysostoses with predominant craniofacial involvement, dysostoses with predominant vertebral involvement, patellar dysostoses, brachydactylies, some disorders with limb hypoplasia-reduction defects, ectrodactyly with and without other manifestations, polydactyly-syndactyly-triphalangism group of disorders, and disorders with defects in joint formation and synostoses. Availability 4 weeks Gene Set Description -

Craniosynostosis Precision Panel Overview Indications Clinical Utility
Craniosynostosis Precision Panel Overview Craniosynostosis is defined as the premature fusion of one or more cranial sutures, often resulting in abnormal head shape. It is a developmental craniofacial anomaly resulting from a primary defect of ossification (primary craniosynostosis) or, more commonly, from a failure of brain growth (secondary craniosynostosis). As well, craniosynostosis can be simple when only one suture fuses prematurely or complex/compound when there is a premature fusion of multiple sutures. Complex craniosynostosis are usually associated with other body deformities. The main morbidity risk is the elevated intracranial pressure and subsequent brain damage. When left untreated, craniosynostosis can cause serious complications such as developmental delay, facial abnormality, sensory, respiratory and neurological dysfunction, eye anomalies and psychosocial disturbances. In approximately 85% of the cases, this disease is isolated and nonsyndromic. Syndromic craniosynostosis usually present with multiorgan complications. The Igenomix Craniosynostosis Precision Panel can be used to make a directed and accurate diagnosis ultimately leading to a better management and prognosis of the disease. It provides a comprehensive analysis of the genes involved in this disease using next-generation sequencing (NGS) to fully understand the spectrum of relevant genes involved. Indications The Igenomix Craniosynostosis Precision Panel is indicated for those patients with a clinical diagnosis or suspicion with or without the following manifestations: ‐ Microcephaly ‐ Scaphocephaly (elongated head) ‐ Anterior plagiocephaly ‐ Brachycephaly ‐ Torticollis ‐ Frontal bossing Clinical Utility The clinical utility of this panel is: - The genetic and molecular confirmation for an accurate clinical diagnosis of a symptomatic patient. - Early initiation of treatment in the form surgical procedures to relieve fused sutures, midface advancement, limited phase of orthodontic treatment and combined 1 orthodontics/orthognathic surgery treatment. -

Identifying the Misshapen Head: Craniosynostosis and Related Disorders Mark S
CLINICAL REPORT Guidance for the Clinician in Rendering Pediatric Care Identifying the Misshapen Head: Craniosynostosis and Related Disorders Mark S. Dias, MD, FAAP, FAANS,a Thomas Samson, MD, FAAP,b Elias B. Rizk, MD, FAAP, FAANS,a Lance S. Governale, MD, FAAP, FAANS,c Joan T. Richtsmeier, PhD,d SECTION ON NEUROLOGIC SURGERY, SECTION ON PLASTIC AND RECONSTRUCTIVE SURGERY Pediatric care providers, pediatricians, pediatric subspecialty physicians, and abstract other health care providers should be able to recognize children with abnormal head shapes that occur as a result of both synostotic and aSection of Pediatric Neurosurgery, Department of Neurosurgery and deformational processes. The purpose of this clinical report is to review the bDivision of Plastic Surgery, Department of Surgery, College of characteristic head shape changes, as well as secondary craniofacial Medicine and dDepartment of Anthropology, College of the Liberal Arts characteristics, that occur in the setting of the various primary and Huck Institutes of the Life Sciences, Pennsylvania State University, State College, Pennsylvania; and cLillian S. Wells Department of craniosynostoses and deformations. As an introduction, the physiology and Neurosurgery, College of Medicine, University of Florida, Gainesville, genetics of skull growth as well as the pathophysiology underlying Florida craniosynostosis are reviewed. This is followed by a description of each type of Clinical reports from the American Academy of Pediatrics benefit from primary craniosynostosis (metopic, unicoronal, bicoronal, sagittal, lambdoid, expertise and resources of liaisons and internal (AAP) and external reviewers. However, clinical reports from the American Academy of and frontosphenoidal) and their resultant head shape changes, with an Pediatrics may not reflect the views of the liaisons or the emphasis on differentiating conditions that require surgical correction from organizations or government agencies that they represent. -

Malformation Syndromes: a Review of Mouse/Human Homology
J Med Genet: first published as 10.1136/jmg.25.7.480 on 1 July 1988. Downloaded from Joalrn(ll of Medical Genetics 1988, 25, 480-487 Malformation syndromes: a review of mouse/human homology ROBIN M WINTER Fromii the Kennetivdy Galton Centre, Clinlicail Research Centre, Northiwick Park Hospital, Harrow, Middlesex HAI 3UJ. SUMMARY The purpose of this paper is to review the known and possible homologies between mouse and human multiple congenital anomaly syndromes. By identifying single gene defects causing similar developmental abnormalities in mouse and man, comparative gene mapping can be carried out, and if the loci in mouse and man are situated in homologous chromosome segments, further molecular studies can be performed to show that the loci are identical. This paper puts forward tentative homologies in the hope that some will be investigated and shown to be true homologies at the molecular level, thus providing mouse models for complex developmental syndromes. The mouse malformation syndromes are reviewed according to their major gene effects. X linked syndromes are reviewed separately because of the greater ease of establishing homology for these conditions. copyright. The purpose of this paper is to review the known even phenotypic similarity would be no guarantee and possible homologies between mouse and human that such genes in man and mouse are homologous". genetic malformation syndromes. Lalley and By identifying single gene defects causing similar following criteria for developmental abnormalities in mouse and man, McKusick' recommend the http://jmg.bmj.com/ identifying gene homologies between species: comparative gene mapping can be carried out, and if (1) Similar nucleotide or amino acid sequence.