Founder Mutations in Tunisia
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Café Au Lait Spots As a Marker of Neuropaediatric Diseases
Mini Review Open Access J Neurol Neurosurg Volume 3 Issue 5 - May 2017 DOI: 10.19080/OAJNN.2017.03.555622 Copyright © All rights are reserved by Francisco Carratalá-Marco Café Au Lait Spots as a Marker of Neuropaediatric Diseases Francisco Carratalá-Marco1*, Rosa María Ruiz-Miralles2, Patricia Andreo-Lillo1, Julia Dolores Miralles-Botella3, Lorena Pastor-Ferrándiz1 and Mercedes Juste-Ruiz2 1Neuropaediatric Unit, University Hospital of San Juan de Alicante, Spain 2Paediatric Department, University Hospital of San Juan de Alicante, Spain 3Dermatology Department, University Hospital of San Juan de Alicante, Spain Submission: March 22, 2017; Published: May 10, 2017 *Corresponding author: Francisco Carratalá-Marco, Neuropaediatric Unit, University Hospital of San Juan de Alicante, Spain, Tel: ; Email: Abstract Introduction: want to know in which The measure,Café au lait the spots presence (CALS) of isolated are shown CALS in representsthe normal a population risk factor forwithout neurological pathological disease. significance, although they could also be criteria for some neurologic syndromes. Unspecific association with general neurologic illnesses has been less frequently described. We Patients and methods: We set up an observational transversal study of cases, patients admitted for neuropaediatric reasons (NPP; n=49) excluding all the patients suffering from neurologic illnesses associated to CALS, and controls, a hospital simultaneously admitted pediatric population for non-neurologic causes (CP; n=101) since October 2012 to January 2013. The data were collected from the clinical reports at admission, and then analyzed by SPSS 22.0 statistical package, and the Stat Calc module of EpiInfo 7.0, following the ethics current rules of the institution for observational studies. -

EFFECTIVE NEBRASKA DEPARTMENT of 01/01/2017 HEALTH and HUMAN SERVICES 173 NAC 1 I TITLE 173 COMMUNICABLE DISEASES CHAPTER 1
EFFECTIVE NEBRASKA DEPARTMENT OF 01/01/2017 HEALTH AND HUMAN SERVICES 173 NAC 1 TITLE 173 COMMUNICABLE DISEASES CHAPTER 1 REPORTING AND CONTROL OF COMMUNICABLE DISEASES TABLE OF CONTENTS SECTION SUBJECT PAGE 1-001 SCOPE AND AUTHORITY 1 1-002 DEFINITIONS 1 1-003 WHO MUST REPORT 2 1-003.01 Healthcare Providers (Physicians and Hospitals) 2 1-003.01A Reporting by PA’s and APRN’s 2 1-003.01B Reporting by Laboratories in lieu of Physicians 3 1-003.01C Reporting by Healthcare Facilities in lieu of Physicians for 3 Healthcare Associated Infections (HAIs) 1-003.02 Laboratories 3 1-003.02A Electronic Ordering of Laboratory Tests 3 1-004 REPORTABLE DISEASES, POISONINGS, AND ORGANISMS: 3 LISTS AND FREQUENCY OF REPORTS 1-004.01 Immediate Reports 4 1-004.01A List of Diseases, Poisonings, and Organisms 4 1-004.01B Clusters, Outbreaks, or Unusual Events, Including Possible 5 Bioterroristic Attacks 1-004.02 Reports Within Seven Days – List of Reportable Diseases, 5 Poisonings, and Organisms 1-004.03 Reporting of Antimicrobial Susceptibility 8 1-004.04 New or Emerging Diseases and Other Syndromes and Exposures – 8 Reporting and Submissions 1-004.04A Criteria 8 1-004.04B Surveillance Mechanism 8 1-004.05 Sexually Transmitted Diseases 9 1-004.06 Healthcare Associated Infections 9 1-005 METHODS OF REPORTING 9 1-005.01 Health Care Providers 9 1-005.01A Immediate Reports of Diseases, Poisonings, and Organisms 9 1-005.01B Immediate Reports of Clusters, Outbreaks, or Unusual Events, 9 Including Possible Bioterroristic Attacks i EFFECTIVE NEBRASKA DEPARTMENT OF -

Connecticut Insurance Coverage for Specialized Formula; New York's “Hannah's Law”
OLR RESEARCH REPORT July 5, 2012 2012-R-0304 CONNECTICUT INSURANCE COVERAGE FOR SPECIALIZED FORMULA; NEW YORK'S “HANNAH'S LAW” By: Janet L. Kaminski Leduc, Senior Legislative Attorney You asked if Connecticut law requires health insurance policies to cover specialized formula used for treating eosinophilic disorder. You also asked for a description of New York’s “Hannah’s Law.” SUMMARY Connecticut insurance law does not explicitly require health insurance policies to cover specialized formulas for treating eosinophilic disorder. However, the law requires specified health insurance policies to cover medically necessary specialized formula for children up to age 12 for treating a disease or condition when the formula is administered under a physician’s direction. Thus, it appears that this would include coverage of specialized formulas for treating eosinophilic disorder. Anyone denied such coverage may appeal to their insurance carrier (internal appeal) or the Insurance Department (external appeal). The explanation of benefits from the carrier describes the appeal process. Anyone with concerns or questions about his or her insurance coverage may contact the Insurance Department’s Consumer Affairs Division at (800) 203-3447 or (860) 297-3900. More information about contacting the department is available at http://www.ct.gov/cid/cwp/view.asp?q=254352. Sandra Norman-Eady, Director Room 5300 Phone (860) 240-8400 Legislative Office Building FAX (860) 240-8881 Connecticut General Assembly Hartford, CT 06106-1591 http://www.cga.ct.gov/olr Office of Legislative Research [email protected] A bill (“Hannah’s Law”) in the New York Assembly would require health insurance policies to cover the cost of enteral formulas for treating eosinophilic esophagitis and related eosinophilic disorders. -

Eye Disease 1 Eye Disease
Eye disease 1 Eye disease Eye disease Classification and external resources [1] MeSH D005128 This is a partial list of human eye diseases and disorders. The World Health Organisation publishes a classification of known diseases and injuries called the International Statistical Classification of Diseases and Related Health Problems or ICD-10. This list uses that classification. H00-H59 Diseases of the eye and adnexa H00-H06 Disorders of eyelid, lacrimal system and orbit • (H00.0) Hordeolum ("stye" or "sty") — a bacterial infection of sebaceous glands of eyelashes • (H00.1) Chalazion — a cyst in the eyelid (usually upper eyelid) • (H01.0) Blepharitis — inflammation of eyelids and eyelashes; characterized by white flaky skin near the eyelashes • (H02.0) Entropion and trichiasis • (H02.1) Ectropion • (H02.2) Lagophthalmos • (H02.3) Blepharochalasis • (H02.4) Ptosis • (H02.6) Xanthelasma of eyelid • (H03.0*) Parasitic infestation of eyelid in diseases classified elsewhere • Dermatitis of eyelid due to Demodex species ( B88.0+ ) • Parasitic infestation of eyelid in: • leishmaniasis ( B55.-+ ) • loiasis ( B74.3+ ) • onchocerciasis ( B73+ ) • phthiriasis ( B85.3+ ) • (H03.1*) Involvement of eyelid in other infectious diseases classified elsewhere • Involvement of eyelid in: • herpesviral (herpes simplex) infection ( B00.5+ ) • leprosy ( A30.-+ ) • molluscum contagiosum ( B08.1+ ) • tuberculosis ( A18.4+ ) • yaws ( A66.-+ ) • zoster ( B02.3+ ) • (H03.8*) Involvement of eyelid in other diseases classified elsewhere • Involvement of eyelid in impetigo -

DIAGNOSIS and TREATMENT of BRUCELLOSIS (Undulant Fever)
DIAGNOSIS AND TREATMENT OF BRUCELLOSIS (Undulant Fever) CHARLES L. HARTSOCK, M.D. Not only the treatment but also the diagnosis of undulant fever are far from being satisfactory, although many types of therapy are being tried and critically evaluated. Because of the tremendous scope of the disease, frequent discussions and reappraisals of our ideas about bru- cellosis will be absolutely essential for some time. Some physicians more or less disregard brucellosis and even scoff at the chronic phase of this new intruder in the realm of human disease. Others are overenthusi- astic and attempt to explain many vague and indefinite problems upon the basis of chronic brucellosis without sufficient evidence. Still other physicians have lost their original enthusiasm and have reverted to the first viewpoint, probably because of the great difficulty in coping with the caprices and vagaries of this disease and the marked uncertainties in diagnosis and treatment. Even though this disease is extremely protean and remarkably bizarre in its manifestations, it is a disease of known causative organism to which the generic term of brucella has been given. The original infection in man was traced to the drinking of goat's milk on the Island of Malta, and for many years this disease was known as Malta fever. Because of the undulating character of the fever with a tendency for remissions and recurrences, it was later called undulant fever which proved to be a very poor description of the febrile reaction in many instances. Brucellosis is the more specific term derived from the organism causing the disease. Three strains of the brucella organism have been isolated and named for their respective hosts: b. -

Analysis of the Tunisian Tax Incentives Regime
Analysis of the Tunisian Tax Incentives Regime March 2013 OECD Paris, France Analysis of the Tunisian Tax Incentives Regime OECD mission, 5-9 November 2012 “…We are working with Tunisia, who joined the Convention on Mutual Administrative Assistance in Tax Matters in July 2012, to review its tax incentives regime and to support its efforts to develop a new investment law.” Remarks by Angel Gurría, OECD Secretary-General, delivered at the Deauville Partnership Meeting of the Finance Ministers in Tokyo, 12 October 2012 1. Executive Summary This analysis of the Tunisian tax incentives regime was conducted by the OECD Tax and Development Programme1 at the request of the Tunisian Ministry of Finance. Following discussions with the government, the OECD agreed to conduct a review of the Tunisian tax incentive system within the framework of the Principles to Enhance the Transparency and Governance of Tax Incentives for Investment in Developing Countries.2 As requested by the Tunisian authorities, the objective of this review was to understand the current system’s bottlenecks and to propose changes to improve efficiency of the system in terms of its ability to mobilise revenue on the one hand and to attract the right kind of investment on the other. The key findings are based on five days of intensive consultations and analysis. Key Findings and Recommendations A comprehensive tax reform effort, including tax policy and tax administration, is critical in the near term to mobilize domestic resources more effectively. The tax reform programme should include, but not be limited to, the development of a new Investment Incentives Code, aimed at transforming the incentives scheme. -

Heat Wave, Raging Fires Take Toll Across Maghreb
12 August 13, 2017 News & Analysis Maghreb Massive loss. A blaze spreading through the hills of a national park north of Tunis. (AFP) Heat wave, raging fires take toll across Maghreb Lamine Ghanmi for Agricultural Production Omar El instigating the fires in the hope of tion that the government would Algerian climatologist Kadi Béhi said nearly 2,000 hectares of drawing benefits from the arson. call them back to work to help put Lamine said: “Indeed, hot weather forest had been lost to fires. “A charcoal mafia had caused the out the fires,” he said. is the beginning of a long period Tunis Algerian Agriculture Minister fires to collect the wood they would Chibani did not comment on a of heat wave because of climate Abdelkader Bouazgui told farmers sell as charcoal during the Eid al- report by the Al Chourouk daily, change. ore than 18,000 hec- during a meeting in the eastern re- Adha feasts,” said Algerian Direc- which quoted “security sources” “The impact of that change tares of the Maghreb’s gion of Jijel that 1,600 fires were re- tor General of Forests Azzedine as saying “the outbreak of the doz- would be swift and brutal. We will forest cover have ported since early July, causing the Sekrane. ens of fires at the same time in the experience extreme weather condi- been lost in about loss of more than 15,000 hectares of He said the fires amounted to north-western forest strip in Tu- tions during summer and winter. 1,700 fire outbreaks forest across northern Algeria. -

C Lif I I I I C Lif I I I I California Immunization Requirements
ClifCaliforn ia IiiImmunization Requirements for School & Child Care Entry Jennifer Sterling San Diego Immunization Partnership www.sdiz.org CtCounty of San Diego, HlthHealth and Human SiServices Agency Immunization Partnership Training Objectives At the end of this training, you will be able to: • List the immunization requirements for child care entry. • Identify acceptable forms of personal immunization records. • Correctly complete the California School Immunization Record (CSIR)/Blue Card. • Create a management system for conditional entrants and exemptions. Why Do We Need Immunizations? • Protect children and families • Prevent illnesses which can cause death and disability • Protect the health of others in the community • Good Public Health Practice Spread of Disease read of Disease –– a a cough, cough, sneezesneeze, or handshake away, or handshake away Vaccine Preventable Diseases in San Diego County Pertussis (Whooping Cough) >604 cases so far in 2010! The worst epidemic in 55 years. • Adults spreading sickness to babies. Nine babies under 3 months of age have died in California this year. • To preven t the sprea d o f Per tuss is, ge t the TDAP vacci ne! Chickenpox • 2009: 69 cases in 10 school districts • 2010: 10 cases in 2 school districts Measles • 2010: Three measles cases in Carlsbad, nine children quarantined • Measles outbreak in San Diego during January and February 2008‐first measles outbreak since 1991! ‐12 cases in 2008 ‐71 peopl e quarantine d Polio is still epidemic in 4 countries! Pertussis – Nearly 50% of the time, infants are infected by their parents Prevention of Disease – SimpleSimple!! Immunizations help prevent the spread of communicable diseases. Cover mouth (w/arm) when coughing or sneezing. -

In the Prevention of Occupational Diseases 94 7.1 Introduction
Report on the current situation in relation to occupational diseases' systems in EU Member States and EFTA/EEA countries, in particular relative to Commission Recommendation 2003/670/EC concerning the European Schedule of Occupational Diseases and gathering of data on relevant related aspects ‘Report on the current situation in relation to occupational diseases’ systems in EU Member States and EFTA/EEA countries, in particular relative to Commission Recommendation 2003/670/EC concerning the European Schedule of Occupational Diseases and gathering of data on relevant related aspects’ Table of Contents 1 Introduction 4 1.1 Foreword .................................................................................................... 4 1.2 The burden of occupational diseases ......................................................... 4 1.3 Recommendation 2003/670/EC .................................................................. 6 1.4 The EU context .......................................................................................... 9 1.5 Information notices on occupational diseases, a guide to diagnosis .................................................................................................. 11 1.6 Objectives of the project ........................................................................... 11 1.7 Methodology and sources ........................................................................ 12 1.8 Structure of the report .............................................................................. 15 2 Developments -

European Conference on Rare Diseases
EUROPEAN CONFERENCE ON RARE DISEASES Luxembourg 21-22 June 2005 EUROPEAN CONFERENCE ON RARE DISEASES Copyright 2005 © Eurordis For more information: www.eurordis.org Webcast of the conference and abstracts: www.rare-luxembourg2005.org TABLE OF CONTENT_3 ------------------------------------------------- ACKNOWLEDGEMENTS AND CREDITS A specialised clinic for Rare Diseases : the RD TABLE OF CONTENTS Outpatient’s Clinic (RDOC) in Italy …………… 48 ------------------------------------------------- ------------------------------------------------- 4 / RARE, BUT EXISTING The organisers particularly wish to thank ACKNOWLEDGEMENTS AND CREDITS 4.1 No code, no name, no existence …………… 49 ------------------------------------------------- the following persons/organisations/companies 4.2 Why do we need to code rare diseases? … 50 PROGRAMME COMMITTEE for their role : ------------------------------------------------- Members of the Programme Committee ……… 6 5 / RESEARCH AND CARE Conference Programme …………………………… 7 …… HER ROYAL HIGHNESS THE GRAND DUCHESS OF LUXEMBOURG Key features of the conference …………………… 12 5.1 Research for Rare Diseases in the EU 54 • Participants ……………………………………… 12 5.2 Fighting the fragmentation of research …… 55 A multi-disciplinary approach ………………… 55 THE EUROPEAN COMMISSION Funding of the conference ……………………… 14 Transfer of academic research towards • ------------------------------------------------- industrial development ………………………… 60 THE GOVERNEMENT OF LUXEMBOURG Speakers ……………………………………………… 16 Strengthening cooperation between academia -
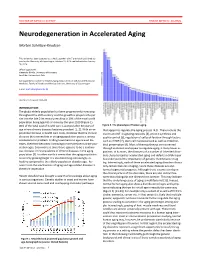
Neurodegeneration in Accelerated Aging
DOCTOR OF MEDICAL SCIENCE DANISH MEDICAL JOURNAL Neurodegeneration in Accelerated Aging Morten Scheibye-Knudsen This review has been accepted as a thesis together with 7 previously published pa- pers by the University of Copenhagen, October 16, 2014 and defended on January 14, 2016 Official opponents: Alexander Bürkle, University of Konstanz Lars Eide, University of Oslo Correspondence: Center for Healthy Aging, Department of Cellular and Molecular Medicine, Faculty of Health and Medical Sciences, University of Copenhagen E-mail: [email protected] Dan Med J 2016;63(11):B5308 INTRODUCTION The global elderly population has been progressively increasing throughout the 20th century and this growth is projected to per- sist into the late 21st century resulting in 20% of the total world population being aged 65 or more by the year 2100 (Figure 1). 80% of the total cost of health care is accrued after 40 years of Figure 2. The phenotype of human aging. age where chronic diseases become prevalent [1, 2]. With an ex- that appear to regulate the aging process [4,5]. These include the ponential increase in health care costs, it follows that the chronic insulin and IGF-1 signaling cascades [4], protein synthesis and diseases that accumulate in an aging population poses a serious quality control [6], regulation of cell proliferation through factors socioeconomic problem. Finding treatments to age related dis- such as mTOR [7], stem cell maintenance 8 as well as mitochon- eases, therefore becomes increasingly more pertinent as the pop- drial preservation [9]. Most of these pathways are conserved ulation ages. Even more so since there appears to be a continu- through evolution and appear to regulate aging in many lower or- ous increase in the prevalence of chronic diseases in the aging ganisms. -

الـجـمهـورية الـتونسـية وزارة الشـؤون الثقافيـة MINISTERE Des AFFAIRES CULTURELLES الـمعهــــــد الوطني للتـــــــراث INSTITUT NATIONAL DU PATRIMOINE
REPUBLIQUE TUNISIENNE الـجـمهـورية الـتونسـية وزارة الشـؤون الثقافيـة MINISTERE des AFFAIRES CULTURELLES الـمعهــــــد الوطني للتـــــــراث INSTITUT NATIONAL DU PATRIMOINE Fiche d’inventaire n° 7/001… 1-Identification de l’élément Nom générique Savoir-faire liés à la poterie modelée des femmes de Sejnane Appellations vernaculaires Fakhār, ṭīn, tamlīs Catégorie Savoir-faire liés à l’artisanat Cadre géographique Sejnane et région de Bizerte La tradition de modeler des poteries selon les techniques usitées à Sejnane perdure dans plusieurs régions de Tunisie, sachant que cette activité fait partie de l’ensemble des tâches domestiques qui incombe à la femme en milieu rural. Les principaux centres de modelage couvrent pratiquement tout le pays : des chaînes montagneuses de la Kroumirie et des Mogods au nord-ouest de la Tunisie, aux villages berbérophones du Djebel Demer dans l’extrême sud, en passant par les bourgs du Cap Bon come Takelsa et Douala, et ceux du Sahel comme El-Jem et Menzel Fersi (ex Sidi Naïja), ainsi 1 que dans la localité de Barrama près de Robaa Siliana. Mais c’est le village de Sejnane et alentours qui sont devenus l’expression la plus achevée de cette tradition.Sejnane est située au nord-ouest du pays, dans la région de Bizerte. Plus précisément, il est implanté sur lemassif montagneux des Mogods, territoire de l'ancienne confédération berbère qui porte le même nom et qui groupe 4 tribus: lesMaalia, Ben Saïdane, S’ḥâbna et M’charga. 2-Description de l’élément Description détaillée L’argile prolifère dans les structures morphologiques du relief, notamment dans les lits d’oued, ce qui facilite son extraction.