The Global Protein-RNA Interaction Map of Epithelial Splicing Regulatory Protein 1 Defines a Post-Transcriptional Program That I
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Ubiquitylome Profiling of Parkin-Null Brain Reveals Dysregulation Of
Neurobiology of Disease 127 (2019) 114–130 Contents lists available at ScienceDirect Neurobiology of Disease journal homepage: www.elsevier.com/locate/ynbdi Ubiquitylome profiling of Parkin-null brain reveals dysregulation of calcium T homeostasis factors ATP1A2, Hippocalcin and GNA11, reflected by altered firing of noradrenergic neurons Key J.a,1, Mueller A.K.b,1, Gispert S.a, Matschke L.b, Wittig I.c, Corti O.d,e,f,g, Münch C.h, ⁎ ⁎ Decher N.b, , Auburger G.a, a Exp. Neurology, Goethe University Medical School, 60590 Frankfurt am Main, Germany b Institute for Physiology and Pathophysiology, Vegetative Physiology and Marburg Center for Mind, Brain and Behavior - MCMBB; Clinic for Neurology, Philipps-University Marburg, 35037 Marburg, Germany c Functional Proteomics, SFB 815 Core Unit, Goethe University Medical School, 60590 Frankfurt am Main, Germany d Institut du Cerveau et de la Moelle épinière, ICM, Paris, F-75013, France e Inserm, U1127, Paris, F-75013, France f CNRS, UMR 7225, Paris, F-75013, France g Sorbonne Universités, Paris, F-75013, France h Institute of Biochemistry II, Goethe University Medical School, 60590 Frankfurt am Main, Germany ARTICLE INFO ABSTRACT Keywords: Parkinson's disease (PD) is the second most frequent neurodegenerative disorder in the old population. Among Parkinson's disease its monogenic variants, a frequent cause is a mutation in the Parkin gene (Prkn). Deficient function of Parkin Mitochondria triggers ubiquitous mitochondrial dysfunction and inflammation in the brain, but it remains unclear howse- Parkin lective neural circuits become vulnerable and finally undergo atrophy. Ubiquitin We attempted to go beyond previous work, mostly done in peripheral tumor cells, which identified protein Calcium targets of Parkin activity, an ubiquitin E3 ligase. -

Dominance of SARS-Cov-2 D614G Variant Explained by the Requirement of COVID-19 for Calcium; Proximate Therapeutic Implication(S) for COVID-19
Cashman DP, J Clin Immunol Immunother 2020, 6: 048 DOI: 10.24966/CIIT-8844/1000048 HSOA Journal of Clinical Immunology and Immunotherapy Research Article Decreased SPCA1 expression causes Hailey-Hailey disease, a rare Dominance of SARS-CoV-2 skin disorder that impairs a cells’ ability to transport Ca2+ (i.e., human ATP2C1 gene). Clinically, the hypocalcemia associated with hospi- D614G Variant Explained by talized COVID-19 patients can effectively impair Ca2+ transport in an analogous manner to the SPCA1 deficiency. Here, the D614G the Requirement of COVID-19 SAR-CoV-2 mutant strain can make use of clinical hypocalcemia like the D104G mutated hPIV-3 virus takes advantage of SPCA1 deficient cells to increase infectivity. These data demonstrate the for Calcium; Proximate critical importance of calcium for effective SARS-CoV-2/COVID-19 infection and how understanding this mechanism can be exploited Therapeutic Implication(s) for for therapeutic gain to stop, or otherwise attenuate virus infection at the earliest steps. Calcium chelation by pharmaceutical EDTA, has COVID-19 been, and can be safely delivered via nebulizer or oral ingestion. Daniel P Cashman M.D.* EDTA treatment is available in outpatient and inpatient settings and can be self-administered during “quarantine” periods. These proven PO Box 841, Los Altos, CA, United States and safe EDTA treatments should effectively disrupt SARS-CoV-2 spread at the earliest step in the virus lifecycle and warrant investi- Abstract gation and optimization to save lives post haste. The current dominance of D614G mutation in the SARS-CoV-2 Keywords: Calcium; Covid-19; Pandemic; SARS-CoV-2; D614G; pandemic implies increased infectivity of the virus S protein that Spike Protein Variant; EDTA; Nebulizer; Treatment; Zinc drives cellular entry which is triggered by binding to the angioten- sin converting enzyme-2 (ACE2) receptor and calcium-dependent Introduction protease-mediated activation. -

Transcriptomic Profiling of Ca Transport Systems During
cells Article Transcriptomic Profiling of Ca2+ Transport Systems during the Formation of the Cerebral Cortex in Mice Alexandre Bouron Genetics and Chemogenomics Lab, Université Grenoble Alpes, CNRS, CEA, INSERM, Bâtiment C3, 17 rue des Martyrs, 38054 Grenoble, France; [email protected] Received: 29 June 2020; Accepted: 24 July 2020; Published: 29 July 2020 Abstract: Cytosolic calcium (Ca2+) transients control key neural processes, including neurogenesis, migration, the polarization and growth of neurons, and the establishment and maintenance of synaptic connections. They are thus involved in the development and formation of the neural system. In this study, a publicly available whole transcriptome sequencing (RNA-Seq) dataset was used to examine the expression of genes coding for putative plasma membrane and organellar Ca2+-transporting proteins (channels, pumps, exchangers, and transporters) during the formation of the cerebral cortex in mice. Four ages were considered: embryonic days 11 (E11), 13 (E13), and 17 (E17), and post-natal day 1 (PN1). This transcriptomic profiling was also combined with live-cell Ca2+ imaging recordings to assess the presence of functional Ca2+ transport systems in E13 neurons. The most important Ca2+ routes of the cortical wall at the onset of corticogenesis (E11–E13) were TACAN, GluK5, nAChR β2, Cav3.1, Orai3, transient receptor potential cation channel subfamily M member 7 (TRPM7) non-mitochondrial Na+/Ca2+ exchanger 2 (NCX2), and the connexins CX43/CX45/CX37. Hence, transient receptor potential cation channel mucolipin subfamily member 1 (TRPML1), transmembrane protein 165 (TMEM165), and Ca2+ “leak” channels are prominent intracellular Ca2+ pathways. The Ca2+ pumps sarco/endoplasmic reticulum Ca2+ ATPase 2 (SERCA2) and plasma membrane Ca2+ ATPase 1 (PMCA1) control the resting basal Ca2+ levels. -

Figure S1. HAEC ROS Production and ML090 NOX5-Inhibition
Figure S1. HAEC ROS production and ML090 NOX5-inhibition. (a) Extracellular H2O2 production in HAEC treated with ML090 at different concentrations and 24 h after being infected with GFP and NOX5-β adenoviruses (MOI 100). **p< 0.01, and ****p< 0.0001 vs control NOX5-β-infected cells (ML090, 0 nM). Results expressed as mean ± SEM. Fold increase vs GFP-infected cells with 0 nM of ML090. n= 6. (b) NOX5-β overexpression and DHE oxidation in HAEC. Representative images from three experiments are shown. Intracellular superoxide anion production of HAEC 24 h after infection with GFP and NOX5-β adenoviruses at different MOIs treated or not with ML090 (10 nM). MOI: Multiplicity of infection. Figure S2. Ontology analysis of HAEC infected with NOX5-β. Ontology analysis shows that the response to unfolded protein is the most relevant. Figure S3. UPR mRNA expression in heart of infarcted transgenic mice. n= 12-13. Results expressed as mean ± SEM. Table S1: Altered gene expression due to NOX5-β expression at 12 h (bold, highlighted in yellow). N12hvsG12h N18hvsG18h N24hvsG24h GeneName GeneDescription TranscriptID logFC p-value logFC p-value logFC p-value family with sequence similarity NM_052966 1.45 1.20E-17 2.44 3.27E-19 2.96 6.24E-21 FAM129A 129. member A DnaJ (Hsp40) homolog. NM_001130182 2.19 9.83E-20 2.94 2.90E-19 3.01 1.68E-19 DNAJA4 subfamily A. member 4 phorbol-12-myristate-13-acetate- NM_021127 0.93 1.84E-12 2.41 1.32E-17 2.69 1.43E-18 PMAIP1 induced protein 1 E2F7 E2F transcription factor 7 NM_203394 0.71 8.35E-11 2.20 2.21E-17 2.48 1.84E-18 DnaJ (Hsp40) homolog. -

Supplementary Table 2
Supplementary Table 2. Differentially Expressed Genes following Sham treatment relative to Untreated Controls Fold Change Accession Name Symbol 3 h 12 h NM_013121 CD28 antigen Cd28 12.82 BG665360 FMS-like tyrosine kinase 1 Flt1 9.63 NM_012701 Adrenergic receptor, beta 1 Adrb1 8.24 0.46 U20796 Nuclear receptor subfamily 1, group D, member 2 Nr1d2 7.22 NM_017116 Calpain 2 Capn2 6.41 BE097282 Guanine nucleotide binding protein, alpha 12 Gna12 6.21 NM_053328 Basic helix-loop-helix domain containing, class B2 Bhlhb2 5.79 NM_053831 Guanylate cyclase 2f Gucy2f 5.71 AW251703 Tumor necrosis factor receptor superfamily, member 12a Tnfrsf12a 5.57 NM_021691 Twist homolog 2 (Drosophila) Twist2 5.42 NM_133550 Fc receptor, IgE, low affinity II, alpha polypeptide Fcer2a 4.93 NM_031120 Signal sequence receptor, gamma Ssr3 4.84 NM_053544 Secreted frizzled-related protein 4 Sfrp4 4.73 NM_053910 Pleckstrin homology, Sec7 and coiled/coil domains 1 Pscd1 4.69 BE113233 Suppressor of cytokine signaling 2 Socs2 4.68 NM_053949 Potassium voltage-gated channel, subfamily H (eag- Kcnh2 4.60 related), member 2 NM_017305 Glutamate cysteine ligase, modifier subunit Gclm 4.59 NM_017309 Protein phospatase 3, regulatory subunit B, alpha Ppp3r1 4.54 isoform,type 1 NM_012765 5-hydroxytryptamine (serotonin) receptor 2C Htr2c 4.46 NM_017218 V-erb-b2 erythroblastic leukemia viral oncogene homolog Erbb3 4.42 3 (avian) AW918369 Zinc finger protein 191 Zfp191 4.38 NM_031034 Guanine nucleotide binding protein, alpha 12 Gna12 4.38 NM_017020 Interleukin 6 receptor Il6r 4.37 AJ002942 -

Human Induced Pluripotent Stem Cell–Derived Podocytes Mature Into Vascularized Glomeruli Upon Experimental Transplantation
BASIC RESEARCH www.jasn.org Human Induced Pluripotent Stem Cell–Derived Podocytes Mature into Vascularized Glomeruli upon Experimental Transplantation † Sazia Sharmin,* Atsuhiro Taguchi,* Yusuke Kaku,* Yasuhiro Yoshimura,* Tomoko Ohmori,* ‡ † ‡ Tetsushi Sakuma, Masashi Mukoyama, Takashi Yamamoto, Hidetake Kurihara,§ and | Ryuichi Nishinakamura* *Department of Kidney Development, Institute of Molecular Embryology and Genetics, and †Department of Nephrology, Faculty of Life Sciences, Kumamoto University, Kumamoto, Japan; ‡Department of Mathematical and Life Sciences, Graduate School of Science, Hiroshima University, Hiroshima, Japan; §Division of Anatomy, Juntendo University School of Medicine, Tokyo, Japan; and |Japan Science and Technology Agency, CREST, Kumamoto, Japan ABSTRACT Glomerular podocytes express proteins, such as nephrin, that constitute the slit diaphragm, thereby contributing to the filtration process in the kidney. Glomerular development has been analyzed mainly in mice, whereas analysis of human kidney development has been minimal because of limited access to embryonic kidneys. We previously reported the induction of three-dimensional primordial glomeruli from human induced pluripotent stem (iPS) cells. Here, using transcription activator–like effector nuclease-mediated homologous recombination, we generated human iPS cell lines that express green fluorescent protein (GFP) in the NPHS1 locus, which encodes nephrin, and we show that GFP expression facilitated accurate visualization of nephrin-positive podocyte formation in -

Transforming Growth Factor ß1-Mediated Functional Inhibition Of
Myelodysplastic Syndromes SUPPLEMENTARY APPENDIX Transforming growth factor 1- mediated functional inhibition of mesenchymal stromal celβls in myelodysplastic syndromes and acute myeloid leukemia Stefanie Geyh, 1* Manuel Rodríguez-Paredes, 1,2 * Paul Jäger, 1 Annemarie Koch, 1 Felix Bormann, 2 Julian Gutekunst, 2 Christoph Zilkens, 3 Ulrich Germing, 1 Guido Kobbe, 1 Frank Lyko, 2 Rainer Haas 1 and Thomas Schroeder 1 1Department of Hematology, Oncology and Clinical Immunology, University of Duesseldorf, Medical Faculty; 2Division of Epigenetics, DKFZ- ZMBH Alliance, German Cancer Research Center, Heidelberg and 3Department of Orthopedic Surgery, University of Duesseldorf, Medical Faculty, Germany *SG and MR-P contributed equally to this work. ©2018 Ferrata Storti Foundation. This is an open-access paper. doi:10.3324/haematol. 2017.186734 Received: December 19, 2017. Accepted: May 14, 2018. Pre-published: May 17, 2018. Correspondence: [email protected] Figure S1 Downregulated genes Downregulated genes Upregulated Figure S1. Heatmaps showing the 50 most upregulated and downregulated genes between the 3 healthy MSC controls and the 9 RCMD-, RAEB- and AML-derived MSC samples. Color scale depicts the rlog-transformed FPKM values for each gene and every sample. Figure S2 Downregulated genes Downregulated genes Upregulated Figure S2. Heatmaps showing the 50 most upregulated and downregulated genes between the 3 healthy MSC controls and the 3 RCMD, RAEB and AML MSC samples, respectively. Color scales depict the rlog-transformed FPKM values for each gene and every sample. Figure S3 A. B. 0.0015 *** ** <-3 -2 0.0010 RCMD RAEB AML -1 0 1 0.0005 Log2FC LTF 2 CCL26/GAPDH INHBB >3 0.0000 TGFB2 y S h D ML M A ealt ll LTF H a EGF 0.003 *** ** INHBB TGFB2 0.002 INHBB IGFBP7 0.001 GDF11 LIF/GAPDH BMP1 0.000 y L th M TNFSF12 l A FGF13 ea ll MDS H a FGF13 0.0015 * TNFSF10 TNFSF10 0.0010 0.0005 SPP1/GAPDH 0.0000 y th l AML ea H all MDS Figure S3. -

Pulmonary Hypertension Remodels the Genomic Fabrics of Major Functional Pathways
Preprints (www.preprints.org) | NOT PEER-REVIEWED | Posted: 21 December 2019 doi:10.20944/preprints201912.0280.v1 Peer-reviewed version available at Genes 2020, 11, 126; doi:10.3390/genes11020126 1 Article 2 Pulmonary Hypertension Remodels the Genomic 3 Fabrics of Major Functional Pathways 4 Rajamma Mathew 1,2, Jing Huang 1, Sanda Iacobas 3 and Dumitru A Iacobas 4,* 5 1 Department of Pediatrics, New York Medical College, Valhalla, NY 10595, U.S.A. 6 2 Department of Physiology, New York Medical College, Valhalla, NY 10595, U.S.A. 7 3 Department of Pathology, New York Medical College, Valhalla, NY 10595, U.S.A. 8 4 Personalized Genomics Laboratory, Center for Computational Systems Biology, Roy G Perry College of 9 Engineering, Prairie View A&M University, Prairie View, TX77446, U.S.A. 10 * Correspondence: [email protected]; Tel.: +1(936)261-9926 11 Abstract: Pulmonary hypertension (PH) is a serious disorder with high morbidity and mortality 12 rate. We analyzed the right ventricular systolic pressure (RVSP), right ventricular hypertrophy 13 (RVH), lung histology and transcriptomes of six weeks old male rats with PH induced by: 1) hypoxia 14 (HO), 2) administration of monocrotaline (CM) or 3) administration of monocrotaline and exposure 15 to hypoxia (HM). The results in PH rats were compared to those in control rats (CO). After four 16 weeks exposure, increased RVSP and RVH, pulmonary arterial wall thickening, and alteration of 17 the lung transcriptome were observed in all PH groups. The HM group exhibited the largest 18 alterations and also neointimal lesions and obliteration of lumen in small arteries. -

Stress-Induced Changes in Primate Prefrontal Profiles of Gene Expression
Molecular Psychiatry (2007) 12, 1089–1102 & 2007 Nature Publishing Group All rights reserved 1359-4184/07 $30.00 www.nature.com/mp ORIGINAL ARTICLE Stress-induced changes in primate prefrontal profiles of gene expression AM Karssen1,6, S Her1,6,7,JZLi2, PD Patel3, F Meng3, WE Bunney Jr4, EG Jones5, SJ Watson3, H Akil3, RM Myers2, AF Schatzberg1 and DM Lyons1 1Department of Psychiatry and Behavioral Sciences, Stanford University, Stanford, CA, USA; 2Stanford Human Genome Center and the Department of Genetics, Stanford University, Stanford, CA, USA; 3Molecular and Behavioral Neuroscience Institute and the Department of Psychiatry, University of Michigan, MI, USA; 4Department of Psychiatry and Human Behavior, University of California, Irvine, CA, USA and 5Center for Neuroscience, University of California, Davis, CA, USA Stressful experiences that consistently increase cortisol levels appear to alter the expression of hundreds of genes in prefrontal limbic brain regions. Here, we investigate this hypothesis in monkeys exposed to intermittent social stress-induced episodes of hypercortisolism or a no-stress control condition. Prefrontal profiles of gene expression compiled from Affymetrix microarray data for monkeys randomized to the no-stress condition were consistent with microarray results published for healthy humans. In monkeys exposed to intermittent social stress, more genes than expected by chance appeared to be differentially expressed in ventromedial prefrontal cortex compared to monkeys not exposed to adult social stress. Most of these stress responsive candidate genes were modestly downregulated, including ubiquitin conjugation enzymes and ligases involved in synaptic plasticity, cell cycle progression and nuclear receptor signaling. Social stress did not affect gene expression beyond that expected by chance in dorsolateral prefrontal cortex or prefrontal white matter. -

Protein Family Members. the GENE.FAMILY
Table 3: Protein family members. The GENE.FAMILY col- umn shows the gene family name defined either by HGNC (superscript `H', http://www.genenames.org/cgi-bin/family_ search) or curated manually by us from Entrez IDs in the NCBI database (superscript `C' for `Custom') that we have identified as corresonding for each ENTITY.ID. The members of each gene fam- ily that are in at least one of our synaptic proteome datasets are shown in IN.SYNAPSE, whereas those not found in any datasets are in the column OUT.SYNAPSE. In some cases the intersection of two HGNC gene families are needed to specify the membership of our protein family; this is indicated by concatenation of the names with an ampersand. ENTITY.ID GENE.FAMILY IN.SYNAPSE OUT.SYNAPSE AC Adenylate cyclasesH ADCY1, ADCY2, ADCY10, ADCY4, ADCY3, ADCY5, ADCY7 ADCY6, ADCY8, ADCY9 actin ActinsH ACTA1, ACTA2, ACTB, ACTC1, ACTG1, ACTG2 ACTN ActininsH ACTN1, ACTN2, ACTN3, ACTN4 AKAP A-kinase anchoring ACBD3, AKAP1, AKAP11, AKAP14, proteinsH AKAP10, AKAP12, AKAP17A, AKAP17BP, AKAP13, AKAP2, AKAP3, AKAP4, AKAP5, AKAP6, AKAP8, CBFA2T3, AKAP7, AKAP9, RAB32 ARFGEF2, CMYA5, EZR, MAP2, MYO7A, MYRIP, NBEA, NF2, SPHKAP, SYNM, WASF1 CaM Endogenous ligands & CALM1, CALM2, EF-hand domain CALM3 containingH CaMKK calcium/calmodulin- CAMKK1, CAMKK2 dependent protein kinase kinaseC CB CalbindinC CALB1, CALB2 CK1 Casein kinase 1C CSNK1A1, CSNK1D, CSNK1E, CSNK1G1, CSNK1G2, CSNK1G3 CRHR Corticotropin releasing CRHR1, CRHR2 hormone receptorsH DAGL Diacylglycerol lipaseC DAGLA, DAGLB DGK Diacylglycerol kinasesH DGKB, -
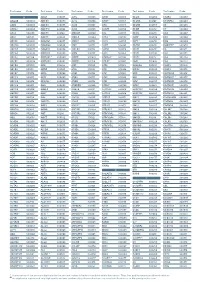
Aagab S00002 Aars S00003 Aars2 S00004 Aass S02483
Test name Code Test name Code Test name Code Test name Code Test name Code Test name Code A ADAR S00053 ALPL S00105 ARSB S00153 BCL10 S02266 C5AR2 S00263 AAGAB S00002 ADCK3 S00054 ALS2 S00106 ARSE * S00154 BCL11A S02167 C5ORF42 S00264 AARS S00003 ADCK4 S00055 ALX3 S00107 ARX S00155 BCL11B S02358 C6 S00265 AARS2 S00004 ADCY10 S02094 ALX4 S00108 ASAH1 S00156 BCOR S00212 C7 S00266 AASS S02483 ADCY3 S02184 AMACR S00109 ASL S00157 BCS1L S00213 C8A S00267 ABAT S02191 ADCY5 S02226 AMELX S02289 ASNS * S02508 BDNF S02509 C8B S00268 ABCA1 S00005 ADGRG1 S00057 AMER1 S00110 ASPA S00158 BDP1 * S00214 C8G S00269 ABCA12 S00006 ADGRG6 S02548 AMH S00111 ASPH S02425 BEAN1 S00215 C8ORF37 S00270 ABCA3 S00007 ADGRV1 S00058 AMHR2 S00112 ASPM S00159 BEST1 S00216 C9 S00271 ABCA4 S00008 ADIPOQ S00059 AMN S00113 ASS1 S00160 BFSP1 S02280 CA2 S00272 ABCA7 S02106 ADIPOR1 * S00060 AMPD1 S02670 ATAD3A * S02196 BFSP2 S00217 CA4 S02303 ABCB11 S00009 ADIPOR2 S00061 AMPD2 S02128 ATCAY S00162 BGN S02633 CA8 S00273 ABCB4 S00010 ADK S02595 AMT S00114 ATF6 S00163 BHLHA9 S00218 CABP2 S00274 ABCB6 S00011 ADNP S02320 ANG S00115 ATIC S02458 BICD2 S00220 CABP4 S00275 ABCB7 S00012 ADSL S00062 ANK1 S00116 ATL1 S00164 BIN1 S00221 CACNA1A S00276 ABCC2 S00013 AFF2 S00063 ANK2 S00117 ATL3 S00165 BLK S00222 CACNA1C * S00277 ABCC6 * S00014 AFG3L2 * S00064 ANKH S00118 ATM S00166 BLM S00223 CACNA1D S00278 ABCC8 S00015 AGA S00065 ANKRD11 * S02140 ATOH7 S02390 BLNK S02281 CACNA1F S00279 ABCC9 S00016 AGBL5 S02452 ANKS6 S00121 ATP13A2 S00168 BLOC1S3 S00224 CACNA1H S00280 ABCD1 * S00017 AGK * -
Human Keratinocyte ATP2C1 Localizes to the Golgi and Controls Golgi Ca2+ Stores
UCSF UC San Francisco Previously Published Works Title Human keratinocyte ATP2C1 localizes to the Golgi and controls Golgi Ca2+ stores Permalink https://escholarship.org/uc/item/5374h7q6 Journal Journal of Investigative Dermatology, 121(4) ISSN 0022-202X Authors Behne, M J Tu, Chia-Ling L Aronchik, I et al. Publication Date 2003-10-01 Peer reviewed eScholarship.org Powered by the California Digital Library University of California ORIGINAL ARTICLE Human Keratinocyte ATP2C1 Localizes to the Golgi and Controls Golgi Ca2 þ Stores Martin J. Behne,Ãw Chia-Ling Tu,z Ida Aronchik,w Ervin Epstein,Ãy Graham Bench,8 Daniel D. Bikle,z Tullio Pozzan,z and Theodora M. MauroÃw ÃDepartment of Dermatology, University of California, San Francisco, California, USA; wDermatology Service and zDepartment of Medicine, Endocrine Unit,VA Medical Center, San Francisco, California, USA; yDepartment of Dermatology, San Francisco General Hospital, San Francisco, California, USA; 8Center for Accelerator Mass Spectrometry, Lawrence Livermore National Laboratory, Livermore, California, USA; zDepartment of Biomedical Sciences and Consiglio Nazionale delle Ricerche (CNR) Center for the Study of Biomembranes, University of Padova, 35121 Padova, Italy Hailey^Hailey disease (MIM16960) is a blistering skin parable to other epithelial cells, Hailey^Hailey disease disease caused by mutations in the Ca2 þ ATPase keratinocyte Golgi Ca2 þ re¢ll is slower, and the maxi- ATP2C1. We found that the abnormal Ca2 þ signaling mum Ca2 þ concentration reached is signi¢cantly lower. seen in Hailey^Hailey disease keratinocytes correlates with These ¢ndings were replicated in vivo, because clinically decreased protein levels of ATP2C1. Human ATP2C1 normal Hailey^Hailey disease epidermis contained low- protein approximated 115 kDa in size.