Scientific Publication of Georeferenced Molecular Data As an Adequate Guide to Delimit the Range of Korean Hynobius Salamanders Through Citizen Science
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

50 CFR Ch. I (10–1–20 Edition) § 16.14
§ 15.41 50 CFR Ch. I (10–1–20 Edition) Species Common name Serinus canaria ............................................................. Common Canary. 1 Note: Permits are still required for this species under part 17 of this chapter. (b) Non-captive-bred species. The list 16.14 Importation of live or dead amphib- in this paragraph includes species of ians or their eggs. non-captive-bred exotic birds and coun- 16.15 Importation of live reptiles or their tries for which importation into the eggs. United States is not prohibited by sec- Subpart C—Permits tion 15.11. The species are grouped tax- onomically by order, and may only be 16.22 Injurious wildlife permits. imported from the approved country, except as provided under a permit Subpart D—Additional Exemptions issued pursuant to subpart C of this 16.32 Importation by Federal agencies. part. 16.33 Importation of natural-history speci- [59 FR 62262, Dec. 2, 1994, as amended at 61 mens. FR 2093, Jan. 24, 1996; 82 FR 16540, Apr. 5, AUTHORITY: 18 U.S.C. 42. 2017] SOURCE: 39 FR 1169, Jan. 4, 1974, unless oth- erwise noted. Subpart E—Qualifying Facilities Breeding Exotic Birds in Captivity Subpart A—Introduction § 15.41 Criteria for including facilities as qualifying for imports. [Re- § 16.1 Purpose of regulations. served] The regulations contained in this part implement the Lacey Act (18 § 15.42 List of foreign qualifying breed- U.S.C. 42). ing facilities. [Reserved] § 16.2 Scope of regulations. Subpart F—List of Prohibited Spe- The provisions of this part are in ad- cies Not Listed in the Appen- dition to, and are not in lieu of, other dices to the Convention regulations of this subchapter B which may require a permit or prescribe addi- § 15.51 Criteria for including species tional restrictions or conditions for the and countries in the prohibited list. -
I Online Supplementary Data – Sexual Size Dimorphism in Salamanders
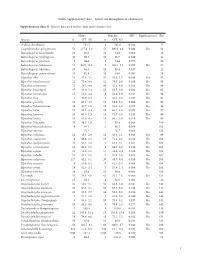
Online Supplementary data – Sexual size dimorphism in salamanders Supplementary data S1. Species data used in this study and references list. Males Females SSD Significant test Ref Species n SVL±SD n SVL±SD Andrias davidianus 2 532.5 8 383.0 -0.280 12 Cryptobranchus alleganiensis 53 277.4±5.2 52 300.9±3.4 0.084 Yes 61 Batrachuperus karlschmidti 10 80.0 10 84.8 0.060 26 Batrachuperus londongensis 20 98.6 10 96.7 -0.019 12 Batrachuperus pinchonii 5 69.6 5 74.6 0.070 26 Batrachuperus taibaiensis 11 92.9±12.1 9 102.1±7.1 0.099 Yes 27 Batrachuperus tibetanus 10 94.5 10 92.8 -0.017 12 Batrachuperus yenyuadensis 10 82.8 10 74.8 -0.096 26 Hynobius abei 24 57.8±2.1 34 55.0±1.2 -0.048 Yes 92 Hynobius amakusaensis 22 75.4±4.8 12 76.5±3.6 0.014 No 93 Hynobius arisanensis 72 54.3±4.8 40 55.2±4.8 0.016 No 94 Hynobius boulengeri 37 83.0±5.4 15 91.5±3.8 0.102 Yes 95 Hynobius formosanus 15 53.0±4.4 8 52.4±3.9 -0.011 No 94 Hynobius fuca 4 50.9±2.8 3 52.8±2.0 0.037 No 94 Hynobius glacialis 12 63.1±4.7 11 58.9±5.2 -0.066 No 94 Hynobius hidamontanus 39 47.7±1.0 15 51.3±1.2 0.075 Yes 96 Hynobius katoi 12 58.4±3.3 10 62.7±1.6 0.073 Yes 97 Hynobius kimurae 20 63.0±1.5 15 72.7±2.0 0.153 Yes 98 Hynobius leechii 70 61.6±4.5 18 66.5±5.9 0.079 Yes 99 Hynobius lichenatus 37 58.5±1.9 2 53.8 -0.080 100 Hynobius maoershanensis 4 86.1 2 80.1 -0.069 101 Hynobius naevius 72.1 76.7 0.063 102 Hynobius nebulosus 14 48.3±2.9 12 50.4±2.1 0.043 Yes 96 Hynobius osumiensis 9 68.4±3.1 15 70.2±3.0 0.026 No 103 Hynobius quelpaertensis 41 52.5±3.8 4 61.3±4.1 0.167 Yes 104 Hynobius -

Salamander Species Listed As Injurious Wildlife Under 50 CFR 16.14 Due to Risk of Salamander Chytrid Fungus Effective January 28, 2016
Salamander Species Listed as Injurious Wildlife Under 50 CFR 16.14 Due to Risk of Salamander Chytrid Fungus Effective January 28, 2016 Effective January 28, 2016, both importation into the United States and interstate transportation between States, the District of Columbia, the Commonwealth of Puerto Rico, or any territory or possession of the United States of any live or dead specimen, including parts, of these 20 genera of salamanders are prohibited, except by permit for zoological, educational, medical, or scientific purposes (in accordance with permit conditions) or by Federal agencies without a permit solely for their own use. This action is necessary to protect the interests of wildlife and wildlife resources from the introduction, establishment, and spread of the chytrid fungus Batrachochytrium salamandrivorans into ecosystems of the United States. The listing includes all species in these 20 genera: Chioglossa, Cynops, Euproctus, Hydromantes, Hynobius, Ichthyosaura, Lissotriton, Neurergus, Notophthalmus, Onychodactylus, Paramesotriton, Plethodon, Pleurodeles, Salamandra, Salamandrella, Salamandrina, Siren, Taricha, Triturus, and Tylototriton The species are: (1) Chioglossa lusitanica (golden striped salamander). (2) Cynops chenggongensis (Chenggong fire-bellied newt). (3) Cynops cyanurus (blue-tailed fire-bellied newt). (4) Cynops ensicauda (sword-tailed newt). (5) Cynops fudingensis (Fuding fire-bellied newt). (6) Cynops glaucus (bluish grey newt, Huilan Rongyuan). (7) Cynops orientalis (Oriental fire belly newt, Oriental fire-bellied newt). (8) Cynops orphicus (no common name). (9) Cynops pyrrhogaster (Japanese newt, Japanese fire-bellied newt). (10) Cynops wolterstorffi (Kunming Lake newt). (11) Euproctus montanus (Corsican brook salamander). (12) Euproctus platycephalus (Sardinian brook salamander). (13) Hydromantes ambrosii (Ambrosi salamander). (14) Hydromantes brunus (limestone salamander). (15) Hydromantes flavus (Mount Albo cave salamander). -

July to December 2019 (Pdf)
2019 Journal Publications July Adelizzi, R. Portmann, J. van Meter, R. (2019). Effect of Individual and Combined Treatments of Pesticide, Fertilizer, and Salt on Growth and Corticosterone Levels of Larval Southern Leopard Frogs (Lithobates sphenocephala). Archives of Environmental Contamination and Toxicology, 77(1), pp.29-39. https://www.ncbi.nlm.nih.gov/pubmed/31020372 Albecker, M. A. McCoy, M. W. (2019). Local adaptation for enhanced salt tolerance reduces non‐ adaptive plasticity caused by osmotic stress. Evolution, Early View. https://onlinelibrary.wiley.com/doi/abs/10.1111/evo.13798 Alvarez, M. D. V. Fernandez, C. Cove, M. V. (2019). Assessing the role of habitat and species interactions in the population decline and detection bias of Neotropical leaf litter frogs in and around La Selva Biological Station, Costa Rica. Neotropical Biology and Conservation 14(2), pp.143– 156, e37526. https://neotropical.pensoft.net/article/37526/list/11/ Amat, F. Rivera, X. Romano, A. Sotgiu, G. (2019). Sexual dimorphism in the endemic Sardinian cave salamander (Atylodes genei). Folia Zoologica, 68(2), p.61-65. https://bioone.org/journals/Folia-Zoologica/volume-68/issue-2/fozo.047.2019/Sexual-dimorphism- in-the-endemic-Sardinian-cave-salamander-Atylodes-genei/10.25225/fozo.047.2019.short Amézquita, A, Suárez, G. Palacios-Rodríguez, P. Beltrán, I. Rodríguez, C. Barrientos, L. S. Daza, J. M. Mazariegos, L. (2019). A new species of Pristimantis (Anura: Craugastoridae) from the cloud forests of Colombian western Andes. Zootaxa, 4648(3). https://www.biotaxa.org/Zootaxa/article/view/zootaxa.4648.3.8 Arrivillaga, C. Oakley, J. Ebiner, S. (2019). Predation of Scinax ruber (Anura: Hylidae) tadpoles by a fishing spider of the genus Thaumisia (Araneae: Pisauridae) in south-east Peru. -

Age Structure and Growth Rates of Two Korean Salamander Species
Animal Cells and Systems Vol. 14, No. 4, December 2010, 315Á322 Age structure and growth rates of two Korean salamander species (Hynobius yangi and Hynobius quelpaertensis) from field populations Jung-Hyun Leea$, Mi-Sook Minb$, Tae-Ho Kimb, Hae-Jun Baekb, Hang Leeb and Daesik Parkc,* aDepartment of Biological Sciences, Kangwon National University, Chuncheon 200-701, Korea; bConservation Genome Resource Bank for Korean Wildlife (CGRB), Research Institute for Veterinary Science, BK21 Program for Veterinary Science and College of Veterinary Medicine, Seoul National University, Seoul 151-742, Korea; cDivision of Science Education, Kangwon National University, Chuncheon 200-701, Korea (Received 20 May 2010; received in revised form 23 August 2010; accepted 24 August 2010) We studied and compared the age structure, body size, and growth rates of field populations of two Korean salamander species (Hynobius yangi and Hynobius quelpaertensis) to elucidate important aspects of basic population dynamics of these two endemic Hynobius species. In both populations, females were sexually mature at three years of age, while H. yangi and H. quelpaertensis males matured at two and three years of age, respectively. Both males and females of H. yangi and H. quelpaertensis attained a maximum age of 11 years and 10 years, respectively. In both species, the snout-vent length (SVL) and body weight (BW) of the females were greater than those of the males. The SVL, BW, and asymptotic SVL of both male and female H. yangi were smaller than those of H. quelpaertensis. The adult growth rates after sexual maturation of male and female H. yangi were lower than those of H. -

DEPARTMENT of the INTERIOR Fish and Wildlife
This document is scheduled to be published in the Federal Register on 01/13/2016 and available online at http://federalregister.gov/a/2016-00452, and on FDsys.gov DEPARTMENT OF THE INTERIOR Fish and Wildlife Service 50 CFR Part 16 RIN 1018–BA77 [Docket No. FWS–HQ–FAC–2015–0005] [FXFR13360900000–156–FF09F14000] Injurious Wildlife Species; Listing Salamanders Due to Risk of Salamander Chytrid Fungus AGENCY: Fish and Wildlife Service, Interior. ACTION: Interim rule; request for comments; notice of availability of economic analysis. 1 SUMMARY: The U.S. Fish and Wildlife Service is amending its regulations under the Lacey Act to add all species of salamanders from 20 genera, of which there are 201 species, to the list of injurious amphibians. With this interim rule, both importation into the United States and interstate transportation between States, the District of Columbia, the Commonwealth of Puerto Rico, or any territory or possession of the United States of any live or dead specimen, including parts, of these 20 genera of salamanders are prohibited, except by permit for zoological, educational, medical, or scientific purposes (in accordance with permit conditions) or by Federal agencies without a permit solely for their own use. This action is necessary to protect the interests of wildlife and wildlife resources from the introduction, establishment, and spread of the chytrid fungus Batrachochytrium salamandrivorans into ecosystems of the United States. The fungus affects salamanders, with lethal effects on many species, and is not yet known to be found in the United States. Because of the devastating effect that we expect the fungus will have on native U.S. -

Title 50 Wildlife and Fisheries Parts 1 to 16
Title 50 Wildlife and Fisheries Parts 1 to 16 Revised as of October 1, 2018 Containing a codification of documents of general applicability and future effect As of October 1, 2018 Published by the Office of the Federal Register National Archives and Records Administration as a Special Edition of the Federal Register VerDate Sep<11>2014 08:08 Nov 27, 2018 Jkt 244234 PO 00000 Frm 00001 Fmt 8091 Sfmt 8091 Y:\SGML\244234.XXX 244234 rmajette on DSKBCKNHB2PROD with CFR U.S. GOVERNMENT OFFICIAL EDITION NOTICE Legal Status and Use of Seals and Logos The seal of the National Archives and Records Administration (NARA) authenticates the Code of Federal Regulations (CFR) as the official codification of Federal regulations established under the Federal Register Act. Under the provisions of 44 U.S.C. 1507, the contents of the CFR, a special edition of the Federal Register, shall be judicially noticed. The CFR is prima facie evidence of the origi- nal documents published in the Federal Register (44 U.S.C. 1510). It is prohibited to use NARA’s official seal and the stylized Code of Federal Regulations logo on any republication of this material without the express, written permission of the Archivist of the United States or the Archivist’s designee. Any person using NARA’s official seals and logos in a manner inconsistent with the provisions of 36 CFR part 1200 is subject to the penalties specified in 18 U.S.C. 506, 701, and 1017. Use of ISBN Prefix This is the Official U.S. Government edition of this publication and is herein identified to certify its authenticity. -

Kim, Jong-Bum; Min, Mi-Sook; Matsui, Masafumi
A new species of lentic breeding Korean salamander of the Title genus Hynobius (Amphibia, Urodela). Author(s) Kim, Jong-Bum; Min, Mi-Sook; Matsui, Masafumi Citation Zoological Science (2003), 20(9): 1163-1169 Issue Date 2003-09 URL http://hdl.handle.net/2433/65044 Right (c) 日本動物学会 / Zoological Society of Japan Type Journal Article Textversion publisher Kyoto University ZOOLOGICAL SCIENCE 20: 1163–1169 (2003) 2003 Zoological Society of Japan A New Species of Lentic Breeding Korean Salamander of the Genus Hynobius (Amphibia, Urodela) Jong-Bum Kim1, 2*, Mi-Sook Min2, 3 and Masafumi Matsui1 1Graduate School of Human and Environmental Studies, Kyoto University, Yoshida Nihonmatsu-cho, Sakyo-ku, Kyoto 606-8501, Japan 2Department of Biological Science,College of Natural Sciences, Inha University, 253, Yonghyun-dong, Nam-ku, Incheon 402-751, Korea 3School of Agricultural Biotechnology and College of Veterinary Medicine, Seoul National University, Suweon 441-744, Korea ABSTRACT—A new species of salamander, Hynobius yangi, is described from Kori (=Hyoam-ri), Busan- shi, southeastern Korea. It is a lentic breeder belonging to the H. nebulosus species group, and has long been confused with another Korean species H. leechii. The new species, however, is genetically substan- tially differentiated from Korean and Japanese relatives. Hynobius yangi is morphologically very similar to H. leechii, but could be differentiated from it by the tail shape, degree of limb separation, shape of vomerine teeth series, shape of egg sac, and dorsal coloration. Key words: cryptic species, Hynobius yangi, Hynobius leechii, Hynobius quelpaertensis, speciation INTRODUCTION SYSTEMATICS In the recent decade, presence of several unique pop- Hynobius yangi sp. -

The Asian Plethodontid Salamander Preserves Historical Genetic Imprints
www.nature.com/scientificreports OPEN The Asian plethodontid salamander preserves historical genetic imprints of recent northern expansion Jong Yoon Jeon1, Ji‑hwa Jung2, Ho Young Suk3, Hang Lee1 & Mi‑Sook Min1* The Korean Peninsula, located at the southern tip of Northeast Asia, has never been covered by ice sheets and was a temperate refugium during the Pleistocene. Karsenia koreana, the sole Asian plethodontid salamander species, occurs only on the southern half of the Korean Peninsula and is thought to have found various climatic refugia. Despite its phylogenetic and biogeographic importance, no population‑level genetic analysis has been performed on this species. Here we study the population genetic structure of K. koreana using mitochondrial and microsatellite loci to understand the recent historical dispersion process that shaped its current distribution. Overall, the genetic distance between populations correlated well with the spatial distance, and the genetic structure among populations showed signs of a unilateral northward expansion from a southernmost refugium population. Given the distinct genetic structure formed among the populations, the level of historical gene fow among populations appears to have been very low. As the estimated efective population size of K. koreana was also small, these results suggest that the small, restricted populations of K. koreana are extremely vulnerable to environmental changes that may require high levels of genetic diversity to cope with. Thus, special management strategies are needed to preserve these remnant populations. Within a geographic area, the distribution of a species refects its historical dispersion and isolation in the context of geological change and Quaternary climate fuctuation1,2. To avoid the advances of Pleistocene ice sheets, many terrestrial animals extirpated or migrated, with some in the northern hemisphere dispersing to the south and occupying sites that served as refugia 3. -

Federal Register/Vol. 81, No. 8/Wednesday, January 13, 2016
1534 Federal Register / Vol. 81, No. 8 / Wednesday, January 13, 2016 / Rules and Regulations If . Then include . ******* (j) the estimated value of the acquisition exceeds $10 million ................ 52.222–24 Pre-award On-site Equal Opportunity Compliance Evalua- tion. (k) the contracting officer requires cost or pricing data for work or serv- 52.215–10 Price Reduction for Defective Certified Cost or Pricing Data. ices exceeding the threshold identified in FAR 15.403–4. 52.215–12 Subcontractor Certified Cost or Pricing Data. ******* [FR Doc. 2016–00475 Filed 1–12–16; 8:45 am] DATES: This interim rule is effective as the offspring or eggs of any of the BILLING CODE 6820–161–P of January 28, 2016. Interested persons foregoing that are injurious to human are invited to submit written comments beings, to the interests of agriculture, on this interim rule on or before March horticulture, or forestry, or to the DEPARTMENT OF THE INTERIOR 14, 2016 wildlife or wildlife resources of the ADDRESSES: You may submit comments United States. Fish and Wildlife Service by any of the following methods: We have determined that salamanders • Federal eRulemaking Portal: http:// that can carry the fungus 50 CFR Part 16 www.regulations.gov. Search for Docket Batrachochytrium salamandrivorans (Bsal) are injurious to wildlife and RIN 1018–BA77 No. FWS–HQ–FAC–2015–0005 and follow the instructions for submitting wildlife resources of the United States. [Docket No. FWS–HQ–FAC–2015–0005; comments. This determination was based on a FXFR13360900000–156–FF09F14000] • Mail, Hand Delivery, or Courier: review of the literature and an Public Comments Processing, Attn: evaluation under the criteria for Injurious Wildlife Species; Listing FWS–HQ–FAC–2015–0005; Division of injuriousness by the Service. -

Contents Contents
Contents Contents Preface 6 Taxonomy and Systematics 8 Biology 12 Metamorphosis 12 The skin 13 Anti-predator behaviour 16 Colours, patterns and colour morphs 21 Thermoregulation 27 Diet 28 Reproduction 29 Natural habitats of tailed amphibians 40 Keeping salamanders and newts 48 Keeping in an aquarium 48 Keeping in a terrarium 50 What to look out for when making a purchase 55 Environmental conditions 55 Population density 64 Handling 67 The right diet 68 Reproduction 78 Sexing 78 Triggers for mating 81 Deposition and development of eggs 84 Development of the larvae 86 Metamorphosis 89 Raising freshly metamorphosed tailed amphibians 90 The right diet for terrestrial juveniles 93 Salamanders, laws and ethics 94 Amphibian decline and keeping salamanders - a contradiction? 94 Laws governing the keeping of newts and salamanders 100 Individual identification and marking 104 Diseases of salamanders and newts 106 How to recognise a sick tailed amphibian 107 2 http://d-nb.info/1059094045 Contents Most common diseases 110 Medical care for diseased tailed amphibians 119 Species accounts of tailed amphibians (Urodela) 120 Salamanders and newts, Salamandridae 120 Calotriton asper (Pyrenean Newt) 120 Cynops cyanurus (Blue-tailed Fire-bellied Newt) 122 Cynops ensicauda (Sword-tailed Newt) 123 Cynops orientalis (Chinese Fire-bellied Newt) 125 Cynops pyrrhogaster (Japanese Fire-bellied Newt) 126 Euproctus platycephalus (Sardinian Newt) 128 Laotriton laoensis (Laos Warty Newt) 130 Lissotriton italicus (Italian Newt) 132 Lissotriton vulgaris (Smooth Newt) 134 Lyciasalamandra billae (Bille's Salamander) 137 Mertensiella caucasica (Caucasian Salamander) 144 Neurergus crocatus (Lake Urmia Newt) 145 Neurergus kaiseri (Luristan Newt) 147 Neurergus strauchii (Anatolia Newt) 149 Notophthalmus viridescens (Red-spotted Newt) 151 Ommatotriton ophryticus (Northern Banded Newt) 153 Ommatotriton vittatus (Southern Banded Newt) 155 Pachytriton sp. -

<I>Hynobius Maoershanensis</I>
HERPETOLOGICAL JOURNAL 18: 129–135, 2008 The karyotype of Hynobius maoershanensis (Urodela: Hynobiidae), a newly described species with rare banding patterns 1,2 2 1 1 3 Qing Li-yan , Wang Yi-ding , Zeng Xiao-mao , Chen Qin- & Hou Mian 1Chengdu Institute of Biology, The Chinese Academy of Sciences, Chengdu, China 2College of Life Sciences, Sichuan Normal University, Chengdu, China 3Continuing Education College of Sichuan Normal University, Chengdu, China The karyotype of Hynobius maoershanensis, a newly described salamander species from mainland China, is described for the first time using Giemsa conventional stain, C-banding and Ag-NORs techniques. All individuals have a diploid number of 56 chromosomes, which is consistent with karyotypes of the pond-type Hynobius group that lives in the lowlands and spawns in small ponds. Several differences distinguish H. maoershanensis from other Hynobius karyotypes: chromosome no. 13 is subtelocentric while in others it is metacentric; there is a prominent and unique dark C-banding pattern encompassing the whole short arm of chromosome no. 13, which can be used as a marker to distinguish this species from other species of Hynobius; multiple C-bands are found only in chromosomes 4, 9 and 13 in this species, much fewer than in other species; NORs are rarely located at the terminal position on the short arm of a large biarmed chromosome (no. 8) in H. maoershanensis, but are commonly observed on the small telocentric microchromosomes (nos. 20, 21 or 23) in other species with 2n=56. All five species from six populations of Hynobius from mainland China can be ecologically attributed to the pond-type group and have 2n=56.