Molecular Characterization of Gastric Epithelial Cells Using Flow Cytometry
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Parietal Cell Hyperplasia Induced by Long-Term Administration of Antacids to Rats
Gut: first published as 10.1136/gut.19.9.798 on 1 September 1978. Downloaded from Gut, 1978, 19, 798-801 Parietal cell hyperplasia induced by long-term administration of antacids to rats G. MAZZACCA1, F. CASCIONE, G. BUDILLON, L. D'AGOSTINO, L. CIMINO, AND C. FEMIANO From the Division of Gastroenterology, 2nd School ofMedicine, University ofNaples, and the Department ofPathology, Institute for Tumoral Diseases, Naples, Italv SUMMARY Suspension of magnesium and aluminum hydroxide (30-60 mEq/24 h) or a comparable volume of water was orally administered by gastric intubation to two groups of 20 male wistar rats each over 60 days. The antacid treatment led to a significant increase in the height (0-464 + 0-02 mm v. 0-318 ± 0 06) and in the volume (472 ± 32 mm3 v. 328 ± 45) of the fundic mucosa ofthe stomach, in the average count of parietal cells per unit area of the mucosa (32-37 ± 1 8 v. 22-3 + 1 6), and in the total parietal cell population of the stomach (53-6 + 3.5 x 106 v. 43-2 + 3.7 x 106). Furthermore fasting serum gastrin concentration was significantly higher in the antacid treated rats (81.2 + 7.4 pg/ml) than in control animals (56-9 ± 6-9 pg/ml). It is known that a feedback mechanism governs the The animals of group B received each time and by relationship between antral gastrin release and the same procedure a comparable volume of water. intraluminal gastric pH (Walsh et al., 1975). During the night each animal was housed in a Furthermore, gastrin exerts a trophic influence on separate cage and the antacid suspension was the the gastric mucosa with increase in the parietal cell only drinking fluid available to the rats of group A. -

Overview of Gastrointestinal Function
Overview of Gastrointestinal Function George N. DeMartino, Ph.D. Department of Physiology University of Texas Southwestern Medical Center Dallas, TX 75390 The gastrointestinal system Functions of the gastrointestinal system • Digestion • Absorption • Secretion • Motility • Immune surveillance and tolerance GI-OP-13 Histology of the GI tract Blood or Lumenal Serosal Side or Mucosal Side Structure of a villus Villus Lamina propria Movement of substances across the epithelial layer Tight junctions X Lumen Blood Apical membrane Basolateral membrane X X transcellular X X paracellular GI-OP-19 Histology of the GI tract Blood or Lumenal Serosal Side or Mucosal Side Motility in the gastrointestinal system Propulsion net movement by peristalsis Mixing for digestion and absorption Separation sphincters Storage decreased pressure GI-OP-42 Intercellular signaling in the gastrointestinal system • Neural • Hormonal • Paracrine GI-OP-10 Neural control of the GI system • Extrinsic nervous system autonomic central nervous system • Intrinsic (enteric) nervous system entirely with the GI system GI-OP-14 The extrinsic nervous system The intrinsic nervous system forms complete functional circuits Sensory neurons Interneurons Motor neurons (excitatory and inhibitory) Parasympathetic nerves regulate functions of the intrinsic nervous system Y Reflex control of gastrointestinal functions Vago-vagal Afferent reflex Salivary Glands Composition of Saliva O Proteins α−amylase lactoferrin lipase RNase lysozyme et al mucus O Electrolyte solution water Na+ , K + - HCO3 -

Expression of Cell Membrane Complement Regulatory Glycoproteins Along the Normal and Diseased Human Gastrointestinal Tract
522 Gut 1998;42:522–529 Expression of cell membrane complement regulatory glycoproteins along the normal and diseased human gastrointestinal tract Gut: first published as 10.1136/gut.42.4.522 on 1 April 1998. Downloaded from A E Berstad, P Brandtzaeg Abstract tract. Indeed, activated complement has been Background/Aims—Uncontrolled comple- detected in lesions of inflammatory bowel ment activation may be of immunopatho- disease,2–4 coeliac disease,5 and recently also in logical importance in inflammatory Helicobacter pylori gastritis.6 diseases of the gastrointestinal tract. Ex- Certain complement activation products, pression of membrane bound factors that notably the C3b fragment and the C5b-7 com- regulate complement activation was there- plex, can on their own—that is, without associ- fore studied in situ. ated antibody—bind to any nearby cell Methods—Frozen tissue specimens were membrane.7 Therefore it is crucial that com- obtained from patients with Helicobacter plement activity is tightly regulated. Mam- pylori gastritis, coeliac disease, Crohn’s malian cells are protected from complement disease, or ulcerative colitis, and from induced damage by a family of cell membrane histologically normal controls. Sections complement regulatory glycoproteins that were examined by immunofluorescence downregulate activation of homologous com- with monoclonal antibodies to protectin plement on their cell surface.8 Protectin (CD59), decay accelerating factor (DAF), (CD59) is broadly distributed on cells of and membrane cofactor protein (MCP). haemopoietic and non-haemopoietic origin79; Results—Protectin and MCP were widely it inhibits the formation of terminal comple- expressed in normal and diseased mu- ment complex by preventing the binding of C9 cosae. -
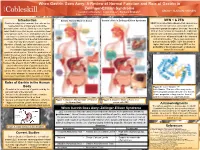
A Review of Normal Function and Role of Gastrin in Zollinger-Ellison
When Gastrin Goes Awry: A Review of Normal Function and Role of Gastrin in Zollinger-Ellison Syndrome Leigha LaTourette1, Janel Cross2, Barbara Brabetz2 Department of Plant and Animal Science1, Department of Natural Sciences and Mathematics2 Introduction Gastrin: Normal Mode of Action Gastrin’s Role in Zollinger-Ellison Syndrome MEN 1 & ZES Gastrin is a digestive hormone that acts on the MEN1 is an inheritable disease that causes over neuroendocrine and parietal cells of the secretion of hormones via tumors on the gastrointestinal tract to ultimately secrete gastric endocrine glands throughout the body.4 Around a acid. Gastrin secretion begins even before food 30% of these tumors are found to be malignant. is consumed, as the mere anticipation of a meal ZES is commonly associated with the MEN1 due can lead to a series of events ending in the to the presence of tumors found in the stomach creation of the gastric acid needed to breakdown and small intestine.4 These gastrointestinal foods. Not only does Gastrin support the tumors caused by MEN 1 give the person a digestion of foods, but it also stimulates growth, higher likelihood of contracting ZES, as the secretion, blood flow, and acts as a defense probability of having pancreatic or duodenal mechanism against bacteria in the tumors is increased.4 gastrointestinal system. When the production of Gastrin becomes overt, major consequences like that of Zollinger Ellison Syndrome (ZES). ZES is an extremely rare disease occurring in people between the ages of 30-60.4 ZES is related to the uncontrolled secretion of gastrin due to the presence of certain pancreatic or duodenal tumors. -

Gastric Fundic Gland Polyps: Can Histology Be Useful to Predict Proton Pump Inhibitors Use?
AG-2018-75 ORIGINAL ARTICLE dx.doi.org/10.1590/S0004-2803.201800000-82 Gastric fundic gland polyps: can histology be useful to predict proton pump inhibitors use? Hugo Leite de Farias BRITO, Cynthia BARROS, Marcelle Vieira FREIRE, Miraldo Nascimento da SILVA FILHO and Tereza Virgínia NASCIMENTO Received 17/6/2018 Accepted 15/10/2018 ABSTRACT – Background – Fundic gland polyps allegedly increased in frequency in recent decades, and had attracted great attention due to possible association with prolonged proton pump inhibitor therapy. Prolonged use of this drug could cause parietal cell hyperplasia, obstruction of glandular lumen and cystic dilation of the gland. Objective – This study aims to analyze clinical and pathological features of fundic gland polyps in patients with and without proton pump inhibitor therapy in a selected population from Brazil. Methods – It was selected a sample of 101 Brazilian patients (78 females and 23 males), from a five years retrospective search of the files from a private pathology laboratory. The patients had an average age of 57 years and we included patients with a histological diagnosis of fundic gland polyp. The clinical data were obtained from their files and all histological slides were reviewed and examined with hematoxylin and eosin (HE) and Giemsa. Results – Information about the use or non-use of proton pump inhibitors (PPI) was obtained in 84 patient files. In 17 cases we could not determine if PPI were used or not. Among those in which the information was available, a positive history of anti-acid therapy was observed in 63 (75.0%) patients. Parietal cell hypertrophy/hyperplasia and parietal cell protrusions were detected in most slides. -

Physiology of the Stomach and Regulation of Gastric Secretions
LECTURE III: Physiology of the Stomach and Regulation of Gastric Secretions EDITING FILE IMPORTANT MALE SLIDES EXTRA FEMALE SLIDES LECTURER’S NOTES 1 PHYSIOLOGY OF THE STOMACH AND REGULATION OF GASTRIC SECRETIONS Lecture Three OBJECTIVES •Functions of stomach. •Gastric secretion. •Mechanism of HCl formation. •Gastric digestive enzymes. •Neural & hormonal control of gastric secretion. •Phases of gastric secretion. •Motor functions of the stomach. •Stomach Emptying. Functional Anatomy of the Stomach: Orad (Reservoir) fundus and upper two orad thirds of the body physiologically Caudad (Antral Pump) lower third of the body plus antrum Caudad Figure 3-1 stomach fundus anatomical body antrum Figure 3-2 ● Gastric mucosa is formed of columnar epithelium that is folded into “pits”. ● The pits are the opening of gastric glands. ● There are several types of gastric glands in the stomach and are distributed differentially in the stomach. Figure 3-3 2 PHYSIOLOGY OF THE STOMACH AND REGULATION OF GASTRIC SECRETIONS Lecture Three Types of Gastric Glands: Mucus secreting glands oxyntic(parietal)glands Pyloric glands (cardiac glands) (most abundant glands) Mucus neck cells Types of cells - Peptic (Chief) cells Many G cells Parietal cells (Oxyntic cells) HCl Gastrin Mucus Pepsinogen secrete Mucus HCO3 IF Mucus Body & fundus Antrum Found in Cardia (above the notch) (below the notch) Proximal 80% of stomach Distal 20% of stomach Structure of a Gastric Oxyntic Gland: ★ HCl is secreted across the parietal cell microvillar membrane and flows out of the intracellular canaliculi into the oxyntic gland lumen. ★ The surface mucous cells line the entire surface of the gastric mucosa and the openings of the cardiac, pyloric, and oxyntic glands. -

Gastrin Receptor (G Protein-Coupled Receptor/Cholecystokinin/Gastric Mucoa) ALAN S
Proc. Nati. Acad. Sci. USA Vol. 89, pp. 3605-3609, April 1992 Biochemistry Expression cloning and characterization of the canine parietal cell gastrin receptor (G protein-coupled receptor/cholecystokinin/gastric mucoa) ALAN S. KOPIN*t, YOUNG-MEE LEE*, EDWARD W. MCBRIDE*, LAURENCE J. MILLER*, MING Lu§, HERBERT Y. LIN¶, LEE F. KOLAKOWSKI, JR. 11, AND MARTIN BEINBORN** *Division of Gastroenterology and GRASP Digestive Disease Center, New England Medical Center, Tufts University School of Medicine, Boston, MA 02111; tGastroenterology Research Unit, Mayo Graduate School of Medicine, Rochester, MN 55905; §Department of Cell Biology, Baylor College of Medicine, Houston, TX 77035; 1Whitehead Institute for Biomedical Research, Cambridge, MA 02142; "1Endocrine Unit, Massachusetts General Hospital, Boston, MA 02114; and **Department of General Pharmacology, Medical School, Hannover, Federal Republic of Germany Communicated by Harvey F. Lodish, January 21, 1992 ABSTRACT Gastrin is an important stimulant of acid tain brain nuclei, has a 1000-fold higher affinity for CCK than secretion by gastric parietal cells and is structurally related to for gastrin. CCK-B/gastrin receptors are found in the brain the peptide hormone cholecystokinin (CCK). The pharmaco- (CCK-B), on smooth muscle cells, and on parietal cells logic properties of the parietal cell gastrin receptor are very (gastrin receptors). Agonist-binding studies on brain mem- similar to the predominant CCK receptor in the brain, CCK-B. branes and parietal cells show a 6- to 10-fold and a 1- to 2-fold Neither the gastrin nor the CCK-B receptor have been cloned higher affinity for CCK than for gastrin, respectively (9). thus far, making it difficult to resolve whether these two These small differences in agonist binding have created receptors are distinct. -

Stomach Tumors Benign Tumors Fundic Gland Polyps Benign
Last updated: 6/11/2020 Prepared by Kurt Schaberg Stomach Tumors Benign Tumors Fundic Gland Polyps Benign. Most common stomach polyp. Hyperplastic expansion of deep oxyntic mucosa with cystically dilated oxyntic glands and foveolar hypoplasia. Parietal cell hyperplasia Usually asymptomatic and incidental. Associated with PPI use Can have CTNNB1 (β-catenin) mutations If numerous (esp. >20) in a young patient, consider a polyposis syndrome, such as FAP. Hyperplastic Polyps Benign. Second most common gastric polyp Elongated, tortuous, hyperplastic foveolar epithelium Cystically dilated glands Inflammatory changes and edema Often eroded at surface. Small, haphazardly distributed smooth muscle Hyperproliferative response to tissue injury. Usually arise in setting of long-standing gastritis Precursor lesion = polypoid foveolar hyperplasia May be hard to differentiate from hamartomatous polyps (e.g., Cronkhite-Canada syndrome) Pyloric Gland adenoma Polypoid proliferation of pyloric-type gastric glands (cuboidal to columnar cells with foamy, ground- glass cytoplasm) and no well-formed apical mucin cap. Often dilated glands. Basal round nuclei. Usually older individuals with atrophic/metaplastic autoimmune gastritis and/or H. pylori Sometimes syndromic (e.g., FAP, GAPPs, etc…) Activating GNAS and/or KRAS mutations and inactivating APC mutations. Can develop high-grade dysplasia → carcinoma Stain with MUC6 Dysplasia Low-grade foveolar-type dysplasia Neoplastic change of gastric epithelium without stromal invasion. Can be gastric/foveolar or intestinal-type (or mixed): Intestinal-type dysplasia: looks like a colonic adenoma with tall columnar cells with hyperchromatic nuclei Gastric/Foveolar-type dysplasia has tubulovillous or serrated fronds lined by cuboidal to columnar cells resembling gastric foveolar cells. Nuclei are round to oval. There is apical neutral mucin. -

CBI 203 Mini Physiology
Introductory Human Physiology ©copyright Emma Jakoi DIGESTIVE SYSTEM -1 Emma Jakoi. Ph.D. LEARNING OBJECTIVES 1. Describe the functional anatomy and role of the digestive system. 2. Describe the production of gastric acid in the stomach. 3. Describe the time course of acid secretion in the fed and fasted states OVERVIEW The digestive system provides nutrients, water, and electrolytes to the cells of the body from the external environment. Food enters the oral cavity and is propelled by muscular contractions through the gastrointestinal (GI) tract moving towards the anus. At various points along the GI tract, acid, digestive enzymes, and buffers are added to facilitate the breakdown of complex foods (such as steak and rice) into simple molecules (such as amino acids, glucose, and fatty acids). These products are then absorbed into the body and delivered to the liver. The various secretions of the GI tract (enzymes, mucus, and water) sum to about 7 liters. This fluid is reabsorbed to prevent dehydration. Unabsorbed nutrients and waste products are eliminated from the body as feces (100 mL – 500mL per day). ANATOMY The digestive system includes the gastrointestinal tract and accessory organs (Fig 1). The gastrointestinal tract is a muscular tube about 5 meters in length. It includes the mouth, pharynx, esophagus, stomach, small intestine and large intestine (colon). Voluntary control occurs at the top and bottom of the tube. Movement through the rest of the gastrointestinal tract is involuntary and unidirectional from mouth to anus. Salivary glands Liver Pancreas Gall bladder Figure 1. Functional regions of the gastrointestinal tract. X marks the sphincters under voluntary control. -

Gastric Secretions
NASPGHAN Physiology Series Gastric Secretions Christine Waasdorp Hurtado, MD, MSCS, FAAP University of Colorado School of Medicine Children’s Hospital Colorado [email protected] Reviewed by Brent Polk, MD and Thomas Sferra, MD H. Pylori (Slides 5-8) H. pylori, flagellated organism, colonize the gastric epithelium of 50% of the world’s population. Complications of infection include gastritis, peptic ulcers, mucosa-associated lymphoid tissue lymphoma (MALT), and gastric cancer. The flagella promote motility in the mucus layer. The organism binds to antigens on gastric epithelial cells, thus preventing mechanical clearance. The organism hydrolyzes urea locally resulting in an increase in gastric pH. Acute infections cause hypochlorhydria due to inhibition of acid secretion. There are three mechanisms involved in acid inhibition; pro-inflammatory cytokine interleukin-1β, suppression of proton pump α-subunit promoter activity and interference in trafficking via tubulovessicles. In chronic infection the stomach may have hypochlorhydria or hyperchlorhydria depending on the severity and location of involvement. Most patients have a pangastritispan gastritis and produce less than normal acid. Twelve percent of infected individuals have an antral dominant infection with inflammation. In antral dominant there is increased acid secretion due to reduced amounts of Somatostatin and increased gastrin. These patients are predisposed to develop a duodenal ulcer. Organism eradication results in normalization of acid, gastrin and Somatostatin. Transmission is by person-to-person contact. Infections are very rare in infants, even if the mother is infected. Re-infection rates are low, but recrudescence (same strain in <12 months) is common. Diagnosis is by one of several tests. Serum H. -

Gastric Acid Secretion in Cholecystokinin-1 Receptor, -2 Receptor, and -1, -2 Receptor Gene Knockout Mice
J Physiol Sci (2009) 59:23–29 DOI 10.1007/s12576-008-0001-y ORIGINAL PAPER Gastric acid secretion in cholecystokinin-1 receptor, -2 receptor, and -1, -2 receptor gene knockout mice Setsuko Kanai Æ Hiroko Hosoya Æ Saeko Akimoto Æ Minoru Ohta Æ Toshimitsu Matsui Æ Soichi Takiguchi Æ Akihiro Funakoshi Æ Kyoko Miyasaka Received: 6 June 2007 / Accepted: 26 September 2008 / Published online: 30 November 2008 Ó The Physiological Society of Japan and Springer 2008 Abstract Gastrin is important for stimulating acid secre- was impaired in mice without CCK-2R. The interaction of tion as well as differentiating gastric mucosal cells via histamine and carbachol was conserved in all genotypes. In cholecystokinin-2 receptors (CCK-2Rs). In turn, CCK acts conclusion, CCK-2R is necessary to respond to carbachol as preferably via CCK-1R to release somatostatin, and well as to produce the maximal acid secretion, while the role somatostatin has been postulated to exhibit a tonic inhibition of CCK-1R in acid secretion is less important. of gastrin bioactivity. The present study was designed to examine the hypothesis that CCK-1R and 2R may act in Keywords Acetylcholine Á CCK Á Knockout Á Mice opposite directions in gastric acid secretion. Having gener- ated CCK-1R(-/-), 2R(-/-), and 1R(-/-)2R(-/-) mice, we examined the regulation of gastric acid secretion in four Introduction genotypes including wild-type mice. Parietal cells possess histamine receptors, muscarinic receptors, and CCK-2Rs. Gastric acid secretion is regulated in a complex manner by Since histamine increases cAMP and carbachol increases neurocrine, endocrine, and paracrine signals. -

THE DIGESTIVE SYSTEM: Gastric Physiology
THE DIGESTIVE SYSTEM: Gastric Physiology Dalay Olson Ph.D Office: Jackson Hall 3-120 Office hours: Friday 11-1 PM Salivary glands Oral cavity Pharynx Lecture 1 Esophagus Upper GI Stomach Liver Lecture 2 Gastric Physiology Pancreas Small intestine Large intestine Lab #1, Lecture 3 & 4 SI, liver, pancreas and LI 3 Compare and contrast the intrinsic, paracrine, endocrine and extrinsic mechanisms of acid secretion. Include secretion released, site of release and intended function. WAYS TO ACTIVATE CELLS IN THE GI TRACT Extrinsic Activation: Activation of the autonomic nervous system • Parasympathetic (Vagus) SNS Vs. PSNS • Sympathetic • Extrinsic mechanisms are long reflexes Intrinsic activation: Activation of the enteric nervous system without input from the CNS. • Mechanoreceptors and chemoreceptors activate ENS ENS activation • Intrinsic mechanisms are short reflexes Paracrine Activation: One cell releases a hormone that has an effect on a neighboring cell. A B • The hormone is released into the interstitial fluid and acts on a nearby cell that expresses the hormone’s receptor. Endocrine Activation: A hormone is released into the blood and has its effects on cells expressing the hormone’s receptor. ACID SECRETION C N You just saw food… S Short Reflex Long Reflex What happens? Your stomach begins producing acid before the food arrives. S T How? O • Parasympathetic stimulation M • Gastrin secreting cells A • Histamine secreting cells C H The release of HCl from the parietal cells relies on the involvement of extrinsic and intrinsic nerves, hormones and paracrine mediators. Mechanisms of acid secretion • Endocrine • Paracrine • Extrinsic (LR) • Intrinsic (SR) SR=Short Reflex LR=Long Reflex Smith, Margaret E., and Dion G.