Does Genetic Structure Reflect Differences in Non-Breeding Movements? a Case Study in Small, Highly Mobile Seabirds
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Birdlife Australia Rarities Committee Unusual Record Report Form
BirdLife Australia Rarities Committee Unusual Record Report Form This form is intended to aid observers in the preparation of a submission for a major rarity in Australia. (It is not a mandatory requirement) Please complete all sections ensuring that you attach all relevant information including any digital images (email to [email protected] or [email protected]). Submissions to BARC should be submitted electronically wherever possible. Full Name: Rob Morris Office Use Address: Phone No: Robert P. Morris, Email: Full Name: Andrew Sutherland (first noticed the second bird) Address: Phone No: Email: Species Name: Broad-billed Prion Scientific Name: Pachyptila vittata Date(s) and time(s) of observation: 11 August 2019 First individual photographed at 12.22 – last bird photographed at 13.11. How long did you watch the bird(s)? c30+ minutes – multiple sightings of 2 birds (possibly 3) and then an additional sighting of 1 bird 20 minutes later whilst travelling, flying past and photographed. First and last date of occurrence: 11 August 2019 Distance to bird: Down to approximately 20-30 m Site Location: SE Tasmania. Approximately 42°50'36.30"S 148°24'46.23"E 22NM ENE of Pirates Bay, Eaglehawk Neck. We went north in an attempt to seek lighter winds and less swell and avoid heading straight into the strong SE winds and southerly swell. Habitat (describe habitat in which the bird was seen): Continental slope waters at a depth of approximately 260 fathoms. Sighting conditions (weather, visibility, light conditions etc.): Weather: Both days were mostly cloudy with occasional periods of bright sunshine. -

DIET and ASPECTS of FAIRY PRIONS BREEDING at SOUTH GEORGIA by P.A
DIET AND ASPECTS OF FAIRY PRIONS BREEDING AT SOUTH GEORGIA By P.A. PRINCE AND P.G. COPESTAKE ABSTRACT A subantarctic population of the Fairy Prion (Pachyprzla turtur) was studied at South Georgia in 1982-83. Full measurements of breeding birds are given, together with details of breeding habitat, the timing of the main breeding cycle events, and chick growth (weight and wing, culmen and tarsus length). Regurgitated food samples showed the diet to be mainly Crustacea (96% by weight), fish and squid comprising the rest. Of crustaceans, Antarctic krill made up 38% of items and 80% by weight. Copepods (four species, mostly Rhincalanus gigas) made up 39% of items but only 4% by weight; amphipods [three species, principally Themisto gaudichaudii made up 22% of items and 16% by weight. Diet and frequency of chick feeding are compared with those of Antarctic Prions and Blue Petrels at the same site; Fairy Prions are essentially intermediate. INTRODUCTION The Fairy Prion (Pachyptila turtur) is one of six members of a genus confined to the temperate and subantarctic regions of the Southern Hemisphere. With the Fulmar Prion (P. crassirostris), it forms the subgenus Pseudoprion. Its main area of breeding distribution is between the Antarctic Polar Front and the Subtropical Convergence. It is widespread in the New Zealand region, from the north of the North Island south to the Antipodes Islands and Macquarie Island, where only about 40 pairs survive (Brothers 1984). Although widespread in the Indian Ocean at the Prince Edward, Crozet and Kerguelen Islands, in the South Atlantic Ocean it is known to breed only on Beauchene Island (Falkland Islands) (Strange 1968, Smith & Prince 1985) and South Georgia (Prince & Croxall 1983). -

Spatial Segregation of Prions and Blue Petrels Is Explained by Differences in Preferred Sea Surface Temper
Animal behaviour Cool, cold or colder? Spatial segregation rsbl.royalsocietypublishing.org of prions and blue petrels is explained by differences in preferred sea surface temperatures Research 1 2 2 2 Cite this article: Quillfeldt P, Cherel Y, Delord Petra Quillfeldt , Yves Cherel , Karine Delord and Henri Weimerkirch K, Weimerkirch H. 2015 Cool, cold or colder? 1Department of Animal Ecology and Systematics, Justus-Liebig-Universita¨t Giessen, 35392 Giessen, Germany Spatial segregation of prions and blue petrels 2Centre d’Etudes Biologiques de Chize´, UMR 7372 CNRS-Universite´ de La Rochelle, 79360 Villiers-en-Bois, France is explained by differences in preferred sea surface temperatures. Biol. Lett. 11: 20141090. The Southern Ocean provides one of the largest environmental gradients on http://dx.doi.org/10.1098/rsbl.2014.1090 Earth that lacks geographical barriers, and small but highly mobile petrels living there may offer fine models of evolution of diversity along environmental gradients. Using geolocation devices, we investigated the winter distribution of closely related petrel species breeding sympatrically in the southern Indian Received: 31 December 2014 Ocean, and applied ecological niche models to compare environmental con- ditions in the habitat used. We show that thin-billed prions (Pachyptila Accepted: 27 March 2015 belcheri), Antarctic prions (Pachyptila desolata) and blue petrels (Halobaena caerulea) from the Kerguelen archipelago in the southern Indian Ocean segre- gate latitudinally, sea surface temperature being the most important variable separating the distribution of the species. Antarctic prions spent the winter Subject Areas: north of the Polar Front in temperate waters, whereas blue petrels were ecology, behaviour found south of the Polar Front in Antarctic waters. -

Master Wildlife List-Peninsula
Antarctic Peninsula Expedition Wildlife List Date Family Species Twitcher List 8-Mar 9-Mar 10-Mar 11-Mar 12-Mar 13-Mar 14-Mar 15-Mar 16-Mar 17-Mar Adelie X X Chinstrap X X X X X X Emperor Penguins Gentoo X X X X X X X Macaroni X X Magellanic X X Unidentifed X X X Black-browed X X X X X Grey-headed X X X X Light-mantled Sooty X X Albatross Northern Royal X X X Southern Royal X X X Wandering X X X Northern X X X X Southern X X X X X X X Giant Petrel Southern (white morph) X X X Unidentified Soft-plumaged Petrel X X X X Gadfly Petrels Soft-plumaged Petrel (Dark) X X Antarctic Petrel Cape Petrel X X X X X X Snow Petrel ? ? Petrels Southern Fulmar X X X X White-chinned Petrel X X X Grey Petrel Great Shearwater X X Shearwaters Sooty Shearwater X X X X Blue Petrel Antarctic Prion X X X X BIRDS Blue Petrels & Prions Fairy Prion Slender-billed Prion X X Unidentified Black-bellied X X X X Grey-backed Storm-petrels Wilson’s X X X X X X X X Unidentified Common Diving-petrels Magellanic Unidentified X X X Antarctic Shag X X X X X X Shags Imperial Shag X X Rock Shag X X Chilean Skua X X South Polar Skua X X X X X X Skuas Subantarctic Skua X X X X X X Unidentified X X Brown-hooded Gull X X Gulls Dolphin Gull X X Kelp Gull X X X X X X X Antarctic X X X X X Terns Arctic South American Blk-crowned Night Heron Blackish Oystercatcher Shorebirds Magellanic Oystercatcher Magellanic Snipe Pale-faced Sheathbill X X X X Other Arnoux's Beaked Blue Cuvier's Beaked Fin Whale Hector's Beaked Humpback X X X Long-finned Pilot Whales Minke X X X X Sei X X X X Southern Bottlenose Southern Right Sperm Beaked (sp.) X X MAMMALS Unidentified Dusky Hourglass X X Orca (Killer Whale) X X Dolphins Peale’s X X Other Unidentified Crabeater X X X X X Leopard X X X X Phocids Southern Elephant X X X Weddell X X X X Antarctic Fur Seal X X X X X Otariids South American Fur Seal South American Sealion OTHER Chilean swallow X X Salp X 8-Mar 9-Mar 10-Mar 11-Mar 12-Mar 13-Mar 14-Mar 15-Mar 16-Mar 17-Mar Twitcher List Date www.oneoceanexpedi,ons.com. -

Part II: Parasite List by Parasite
Surveillance Vol.25 Special Issue 1998 Parasites of Birds in New Zealand Part II: Parasite list by parasite Ectoparasite Host Ectoparasite _- Host Feather mites Antrlges sp ~ continued Allopt~ssp Gull, Red-billed Chaff i11 ch Gannet, Australasian Creeper, Brown Shearwater, Flesh-footed Morepork Booby, Masked (Blue-faced) Sparrow, House Booby, Brown Saddleback, South Island Plover, Shore Tu i Sparrow, Hedge Alloptes hisetatus Tern, Caspian Canary Alloptes phaetontis minor Tropicbird, Red-tailed Quail, Brown Petrel, White-naped Blackbird Thrush, Song Allopres phaeton tis simplex Tropicbird, Red-tailed Andgopsis paxserinus Myna, Common Starling Alloptes stercorurii Skua, Arctic Anheniialges sp Fantail, South Island Ana1ge.r sp Myna, Common Fantail, North Island Skylark Greenfinch Brephosceles sp Dotterel, New Zealand Rook Mollymawk, Buller’s Parakeet, Yellow-crowned Petrel, Black-bellied Storm Yellowhammer Petrel, Northern Giant 26 Surveillance Special Issue Surveillance Vol.25 Special Issue 1998 Parasites of Birds in New Zealand Host Ectoparasite I Weka Prion, Broad-hi1 led Kea Petrel. Cook's Kakapo Pe tre I, M age11 ta (Chatham Pigeon. Rock Island Taiko) Fowl. Domestic Shearwater. Flesh-footed Shearwater, Fluttering Weka Wcka I lot t c re I. Banded Pheasant, Ring-necked Pctrcl. White-naped Pipn. New Zealand Petrel . W hi te-naped Dotterel, New Zealand Pukeko Shearwater. Sooty Rail, Banded Ovstercatcher. South Island Greenfinch Sparrow. HOLIX Pkd Stilt. Australasian Pied Kokako. South Island Stilt. Blach Weka Plover, Shore Kakapo S iIvere ye Pigeon, New Zealand Petrel. White-napcd Pukeko Petrel. Cook's Rail. Banded Petrel. Black-winged Shag. Little Petrel. Pycroft's Shag. Little Black (Little Shag, Campbell Island Black Cormorant) Shag, Stewart Island. -

Preparations for the Eradication of Mice from Gough Island: Results of Bait Acceptance Trials Above Ground and Around Cave Systems
Cuthbert, R.J.; P. Visser, H. Louw, K. Rexer-Huber, G. Parker, and P.G. Ryan. Preparations for the eradication of mice from Gough Island: results of bait acceptance trials above ground and around cave systems Preparations for the eradication of mice from Gough Island: results of bait acceptance trials above ground and around cave systems R. J. Cuthbert1, P. Visser1, H. Louw1, K. Rexer-Huber1, G. Parker1, and P. G. Ryan2 1Royal Society for the Protection of Birds, The Lodge, Sandy, Bedfordshire, SG19 2DL, United Kingdom. <[email protected]>. 2DST/NRF Centre of Excellence at the Percy FitzPatrick Institute, University of Cape Town, Rondebosch 7701, South Africa. Abstract Gough Island, Tristan da Cunha, is a United Kingdom Overseas Territory, supports globally important seabird colonies, has many endemic plant, invertebrate and bird taxa, and is recognised as a World Heritage Site. A key threat to the biodiversity of Gough Island is predation by the introduced house mouse (Mus musculus), as a result of which two bird species are listed as Critically Endangered. Eradicating mice from Gough Island is thus an urgent conservation priority. However, the higher failure rate of mouse versus rat eradications, and smaller size of islands that have been successfully cleared of mice, means that trials on bait acceptance are required to convince funding agencies that an attempted eradication of mice from Gough is likely to succeed. In this study, trials of bait acceptance were undertaken above ground and around cave systems that are potential refuges for mice during an aerial application of bait. Four trials were undertaken during winter, with rhodamine-dyed, non-toxic bait spread by hand at 16 kg/ha over 2.56 ha centred above cave systems in Trials 1-3 and over 20.7 ha and two caves in Trial 4. -

Densities of Antarctic Seabirds at Sea and the Presence of the Krill Eupha Usia Superba
DENSITIES OF ANTARCTIC SEABIRDS AT SEA AND THE PRESENCE OF THE KRILL EUPHA USIA SUPERBA BRYAN S. OBST Departmentof Biology,University of California,Los Angeles, California 90024 USA ABSTRACT.--Theantarctic krill Euphausiasuperba forms abundant,well-organized schools in the watersoff the AntarcticPeninsula. Mean avian densityis 2.6 timesgreater in waters where krill schoolsare present than in waters without krill schools.Seabird density is a good predictorof the presenceof krill. Seabirddensity did not correlatewith krill density or krill schooldepth. Disoriented krill routinely were observedswimming near the surface above submergedschools, providing potential prey for surface-feedingbirds. Responsesof seabird speciesto the distribution of krill schoolsvaried. The small to me- dium-sizeprocellariiform species were the best indicatorsof krill schools;large procellari- iforms and coastalspecies were poor indicators.Pygoscelis penguins occurredat high den- sitiesonly in the presenceof krill schools.These responses are consistentwith the constraints imposedby the metabolicrequirements and reproductivestrategies of eachof thesegroups. Krill schoolswere detectednear the seasurface throughout the day. Correlationsbetween seabirddensity and the presenceof krill during daylight hourssuggest that diurnal foraging is important to the seabirdsof this region. Received19 December1983, accepted4 December 1984. RELATIVELY little is known about the factors birds depend on directly or indirectly for food influencing the distribution of seabirdsin the (Haury et al. 1978). These observationssuggest marine habitat. The past decade has produced that relatively small-scalephenomena, such as a number of studiesattempting to correlatepat- local concentrationsof prey, may be of major terns of avian abundance and distribution with importance in determining the patterns of sea- physical featuresof the oceansuch as currents bird distribution within the broad limits set by and convergences,water masses,and temper- featuresof the physical ocean. -
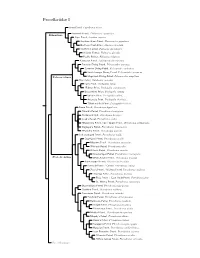
Procellariidae Species Tree
Procellariidae I Snow Petrel, Pagodroma nivea Antarctic Petrel, Thalassoica antarctica Fulmarinae Cape Petrel, Daption capense Southern Giant-Petrel, Macronectes giganteus Northern Giant-Petrel, Macronectes halli Southern Fulmar, Fulmarus glacialoides Atlantic Fulmar, Fulmarus glacialis Pacific Fulmar, Fulmarus rodgersii Kerguelen Petrel, Aphrodroma brevirostris Peruvian Diving-Petrel, Pelecanoides garnotii Common Diving-Petrel, Pelecanoides urinatrix South Georgia Diving-Petrel, Pelecanoides georgicus Pelecanoidinae Magellanic Diving-Petrel, Pelecanoides magellani Blue Petrel, Halobaena caerulea Fairy Prion, Pachyptila turtur ?Fulmar Prion, Pachyptila crassirostris Broad-billed Prion, Pachyptila vittata Salvin’s Prion, Pachyptila salvini Antarctic Prion, Pachyptila desolata ?Slender-billed Prion, Pachyptila belcheri Bonin Petrel, Pterodroma hypoleuca ?Gould’s Petrel, Pterodroma leucoptera ?Collared Petrel, Pterodroma brevipes Cook’s Petrel, Pterodroma cookii ?Masatierra Petrel / De Filippi’s Petrel, Pterodroma defilippiana Stejneger’s Petrel, Pterodroma longirostris ?Pycroft’s Petrel, Pterodroma pycrofti Soft-plumaged Petrel, Pterodroma mollis Gray-faced Petrel, Pterodroma gouldi Magenta Petrel, Pterodroma magentae ?Phoenix Petrel, Pterodroma alba Atlantic Petrel, Pterodroma incerta Great-winged Petrel, Pterodroma macroptera Pterodrominae White-headed Petrel, Pterodroma lessonii Black-capped Petrel, Pterodroma hasitata Bermuda Petrel / Cahow, Pterodroma cahow Zino’s Petrel / Madeira Petrel, Pterodroma madeira Desertas Petrel, Pterodroma -

Conservation Advice Pachyptila Tutur Subantarctica
THREATENED SPECIES SCIENTIFIC COMMITTEE Established under the Environment Protection and Biodiversity Conservation Act 1999 The Minister’s delegate approved this conservation advice on 01/10/2015 Conservation Advice Pachyptila tutur subantarctica fairy prion (southern) Conservation Status Pachyptila tutur subantarctica (fairy prion (southern)) is listed as Vulnerable under the Environment Protection and Biodiversity Conservation Act 1999 (Cwlth) (EPBC Act). The species is eligible for listing as Vulnerable as, prior to the commencement of the EPBC Act, it was listed as Vulnerable under Schedule 1 of the Endangered Species Protection Act 1992 (Cwlth). The main factor that is the cause of the species being eligible for listing in the Vulnerable category is its small population size (250-1000 mature individuals). The Action Plan for Australian Birds 2010 considered the Australian breeding population of the fairy prion (southern) as Endangered under Criterion D (Australian breeding population is <250 mature individuals) (Garnett et al., 2011). The Threatened Species Scientific Committee are using the findings of Garnett et al., (2011) to consider whether reassessment of the conservation status of each of threatened birds listed under the EPBC Act is required. Description The fairy prion is the smallest of the Australian prions, with a wingspan of about 56 cm. They have a short narrow bill with a strong hook narrowly separated from the nasal tubes. The fairy prion is grey-blue above, with a darker grey crown, and grey eye stripe. The eyes are dark and highlighted above and below the grey eye stripe by white plumage. The tail is triangular and has a thick dark bank at the tip (Pizzey & Knight 1999). -

Niche Switching and Leapfrog Foraging: Movement Ecology of Sympatric Petrels During the Early Breeding Season
Quillfeldt et al. Movement Ecology (2020) 8:23 https://doi.org/10.1186/s40462-020-00212-y RESEARCH Open Access Niche switching and leapfrog foraging: movement ecology of sympatric petrels during the early breeding season Petra Quillfeldt1* , Henri Weimerskirch2, Karine Delord2 and Yves Cherel2 Abstract Background: The timing of events in the early part of the breeding season is crucially important for successful reproduction. Long-lived animals that migrate large distances independently of each other meet at the breeding sites to re-establish their pair bonds and coordinate their breeding duties with their partners. Methods: Using miniature light-geolocation and immersion data together with blood stable isotopes, we studied the early breeding season in Thin-billed prions Pachyptila belcheri, Antarctic prions P. desolata and Blue petrels Halobaena caerulea breeding at Kerguelen Islands in the Indian Ocean. These three species exhibit differences in their winter habitat and timing of migration, moult and breeding. We hypothesised that these differences would influence their behaviour during the early breeding season. Results: In line with our hypothesis, we found clear differences not only in the timing of colony attendance, but also in the time budgets while at sea and in habitat use. Both early breeding Blue petrels and late breeding Antarctic prions spent about 8 h per day in flight and 15 h foraging. In comparison, Thin-billed prions, which breed in mid- summer, spent less time (5 h daily) in flight and more time (18 h daily) foraging, thus maximizing the time spent foraging during the longest daylight days of the year. While the ecological habitat parameters (sea temperature, wind, productivity) of Thin-billed prions and Blue petrels were relatively stable throughout the year, Antarctic prions showed clear niche switching, caused by leapfrogging between the northernmost winter distribution to the southernmost distribution during the early breeding season. -

Order PROCELLARIIFORMES: Albatrosses, Petrels, Prions and Shearwaters Family PROCELLARIIDAE Leach
Text extracted from Gill B.J.; Bell, B.D.; Chambers, G.K.; Medway, D.G.; Palma, R.L.; Scofield, R.P.; Tennyson, A.J.D.; Worthy, T.H. 2010. Checklist of the birds of New Zealand, Norfolk and Macquarie Islands, and the Ross Dependency, Antarctica. 4th edition. Wellington, Te Papa Press and Ornithological Society of New Zealand. Pages 64, 78-79, 98-99 & 101-102. Order PROCELLARIIFORMES: Albatrosses, Petrels, Prions and Shearwaters Checklist Committee (1990) recognised three families within the Procellariiformes, however, four families are recognised here, with the reinstatement of Pelecanoididae, following many other recent authorities (e.g. Marchant & Higgins 1990, del Hoyo et al. 1992, Viot et al. 1993, Warham 1996: 484, Nunn & Stanley 1998, Dickinson 2003, Brooke 2004, Onley & Scofield 2007). The relationships of the families within the Procellariiformes are debated (e.g. Sibley & Alquist 1990, Christidis & Boles 1994, Nunn & Stanley 1998, Livezey & Zusi 2001, Kennedy & Page 2002, Rheindt & Austin 2005), so a traditional arrangement (Jouanin & Mougin 1979, Marchant & Higgins 1990, Warham 1990, del Hoyo et al. 1992, Warham 1996: 505, Dickinson 2003, Brooke 2004) has been adopted. The taxonomic recommendations (based on molecular analysis) on the Procellariiformes of Penhallurick & Wink (2004) have been heavily criticised (Rheindt & Austin 2005) and have seldom been followed here. Family PROCELLARIIDAE Leach: Fulmars, Petrels, Prions and Shearwaters Procellariidae Leach, 1820: Eleventh room. In Synopsis Contents British Museum 17th Edition, London: 68 – Type genus Procellaria Linnaeus, 1758. Subfamilies Procellariinae and Fulmarinae and shearwater subgenera Ardenna, Thyellodroma and Puffinus (as recognised by Checklist Committee 1990) are not accepted here given the lack of agreement about to which subgenera some species should be assigned (e.g. -

Falkland Islands Species List
Falkland Islands Species List Day Common Name Scientific Name x 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 1 BIRDS* 2 DUCKS, GEESE, & WATERFOWL Anseriformes - Anatidae 3 Black-necked Swan Cygnus melancoryphus 4 Coscoroba Swan Coscoroba coscoroba 5 Upland Goose Chloephaga picta 6 Kelp Goose Chloephaga hybrida 7 Ruddy-headed Goose Chloephaga rubidiceps 8 Flying Steamer-Duck Tachyeres patachonicus 9 Falkland Steamer-Duck Tachyeres brachypterus 10 Crested Duck Lophonetta specularioides 11 Chiloe Wigeon Anas sibilatrix 12 Mallard Anas platyrhynchos 13 Cinnamon Teal Anas cyanoptera 14 Yellow-billed Pintail Anas georgica 15 Silver Teal Anas versicolor 16 Yellow-billed Teal Anas flavirostris 17 GREBES Podicipediformes - Podicipedidae 18 White-tufted Grebe Rollandia rolland 19 Silvery Grebe Podiceps occipitalis 20 PENGUINS Sphenisciformes - Spheniscidae 21 King Penguin Aptenodytes patagonicus 22 Gentoo Penguin Pygoscelis papua Cheesemans' Ecology Safaris Species List Updated: April 2017 Page 1 of 11 Day Common Name Scientific Name x 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 23 Magellanic Penguin Spheniscus magellanicus 24 Macaroni Penguin Eudyptes chrysolophus 25 Southern Rockhopper Penguin Eudyptes chrysocome chrysocome 26 ALBATROSSES Procellariiformes - Diomedeidae 27 Gray-headed Albatross Thalassarche chrysostoma 28 Black-browed Albatross Thalassarche melanophris 29 Royal Albatross (Southern) Diomedea epomophora epomophora 30 Royal Albatross (Northern) Diomedea epomophora sanfordi 31 Wandering Albatross (Snowy) Diomedea exulans exulans 32 Wandering