Applied Regression Analysis Course Notes for STAT 378/502
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Computer-Assisted Data Treatment in Analytical Chemometrics III. Data Transformation
Computer-Assisted Data Treatment in Analytical Chemometrics III. Data Transformation aM. MELOUN and bJ. MILITKÝ department of Analytical Chemistry, Faculty of Chemical Technology, University Pardubice, CZ-532 10 Pardubice ъDepartment of Textile Materials, Technical University, CZ-461 17 Liberec Received 30 April 1993 In trace analysis an exploratory data analysis (EDA) often finds that the sample distribution is systematically skewed or does not prove a sample homogeneity. Under such circumstances the original data should often be transformed. The power simple transformation and the Box—Cox transformation improves a sample symmetry and also makes stabilization of variance. The Hines— Hines selection graph and the plot of logarithm of the maximum likelihood function enables to find an optimum transformation parameter. Procedure of data transformation in the univariate data analysis is illustrated on quantitative determination of copper traces in kaolin raw. When exploratory data analysis shows that the i) Transformation for variance stabilization implies sample distribution strongly differs from the normal ascertaining the transformation у = g(x) in which the one, we are faced with the problem of how to analyze variance cf(y) is constant. If the variance of the origi the data. Raw data may require re-expression to nal variable x is a function of the type cf(x) = ^(x), produce an informative display, effective summary, the variance cr(y) may be expressed by or a straightforward analysis [1—10]. We may need to change not only the units in which the data are 2 fdg(x)f stated, but also the basic scale of the measurement. <У2(У)= -^ /i(x)=C (1) To change the shape of a data distribution, we must I dx J do more than change the origin and/or unit of mea surement. -
Appendix: Statistical Tables
Appendix: Statistical Tables This appendix contains statistical tables of the common sampling distributions used in statistical inference. See Chapter 4 for examples of their use. CUMULATIVE DISTRIBUTION FUNCTION OF THE STANDARD NORMAL DISTRIBUTION Table A.1 shows the probability, F(z) that a standard normal random variable is less than the value z. For example, the probability is 0.9986 that a standard normal random variable is less than 3. CHI-SQUARE DISTRIBUTION For a given probabilities α, Table A.2 shows the values of the chi-square distribution. For example, the probability is 0.05 that a chi-square random variable with 10 degrees of freedom is greater than 18.31. STUDENT T-DISTRIBUTION For a given probability α, Table A.3 shows the values of the student t-distribution. For example, the probability is 0.05 that a student t random variable with 10 degrees of freedom is greater than 1.812. 227 228 Table A.1 Cumulative distribution function of the standard normal distribution zF(z) zF(z) ZF(z) zF(z) 0 0.5 0.32 0.625516 0.64 0.738914 0.96 0.831472 0.01 0.503989 0.33 0.6293 0.65 0.742154 0.97 0.833977 0.02 0.507978 0.34 0.633072 0.66 0.745373 0.98 0.836457 0.03 0.511967 0.35 0.636831 0.67 0.748571 0.99 0.838913 0.04 0.515953 0.36 0.640576 0.68 0.751748 1 0.841345 0.05 0.519939 0.37 0.644309 0.69 0.754903 1.01 0.843752 0.06 0.523922 0.38 0.648027 0.7 0.758036 1.02 0.846136 0.07 0.527903 0.39 0.651732 0.71 0.761148 1.03 0.848495 0.08 0.531881 0.4 0.655422 0.72 0.764238 1.04 0.85083 0.09 0.535856 0.41 0.659097 0.73 0.767305 1.05 0.85314 -

UNIT-III: Correlation and Regression
BUSINESS STATISTICS BBA UNIT-III 2ND SEM UNIT-III: Correlation and Regression: Meaning of correlation, types of correlation – positive and negative correlation, simple, partial and multiple correlation, methods of studying correlation; scatter diagram, graphic and direct method; properties of correlation co-efficient, rank correlation, coefficient of determination, lines of regression, co-efficient of regression, standard error of estimate. Correlation Correlation is used to test relationships between quantitative variables or categorical variables. In other words, it’s a measure of how things are related. The study of how variables are correlated is called correlation analysis. Some examples of data that have a high correlation: Your caloric intake and your weight. Your eye color and your relatives’ eye colors. Some examples of data that have a low correlation (or none at all): A dog’s name and the type of dog biscuit they prefer. The cost of a car wash and how long it takes to buy a soda inside the station. Correlations are useful because if you can find out what relationship variables have, you can make predictions about future behavior. Knowing what the future holds is very important in the social sciences like government and healthcare. Businesses also use these statistics for budgets and business plans. Scatter Diagram A scatter diagram is a diagram that shows the values of two variables X and Y, along with the way in which these two variables relate to each other. The values of variable X are given along the horizontal axis, with the values of the variable Y given on the vertical axis. -

1. a Dependent Variable Is Also Known As A(N) ___
1. A dependent variable is also known as a(n) _____. a. explanatory variable b. control variable c. predictor variable d. response variable ANSWER: d RATIONALE: FEEDBACK: A dependent variable is known as a response variable. POINTS: 1 DIFFICULTY: Easy NATIONAL STANDARDS: United States - BUSPROG: Analytic TOPICS: Definition of the Simple Regression Model KEYWORDS: Bloom’s: Knowledge 2. If a change in variable x causes a change in variable y, variable x is called the _____. a. dependent variable b. explained variable c. explanatory variable d. response variable ANSWER: c RATIONALE: FEEDBACK: If a change in variable x causes a change in variable y, variable x is called the independent variable or the explanatory variable. POINTS: 1 DIFFICULTY: Easy NATIONAL STANDARDS: United States - BUSPROG: Analytic TOPICS: Definition of the Simple Regression Model KEYWORDS: Bloom’s: Comprehension 3. In the equation is the _____. a. dependent variable b. independent variable c. slope parameter d. intercept parameter ANSWER: d RATIONALE: FEEDBACK: In the equation is the intercept parameter. POINTS: 1 DIFFICULTY: Easy NATIONAL STANDARDS: United States - BUSPROG: Analytic TOPICS: Definition of the Simple Regression Model KEYWORDS: Bloom’s: Knowledge 4. In the equation , what is the estimated value of ? a. b. Cengage Learning Testing, Powered by Cognero Page 1 c. d. ANSWER: a RATIONALE: FEEDBACK: The estimated value of is . POINTS: 1 DIFFICULTY: Easy NATIONAL STANDARDS: United States - BUSPROG: Analytic TOPICS: Deriving the Ordinary Least Squares Estimates KEYWORDS: Bloom’s: Knowledge 5. In the equation , c denotes consumption and i denotes income. What is the residual for the 5th observation if =$500 and =$475? a. -

Systematic Evaluation and Optimization of Outbreak-Detection Algorithms Based on Labeled Epidemiological Surveillance Data
Universit¨atOsnabr¨uck Fachbereich Humanwissenschaften Institute of Cognitive Science Masterthesis Systematic Evaluation and Optimization of Outbreak-Detection Algorithms Based on Labeled Epidemiological Surveillance Data R¨udigerBusche 968684 Master's Program Cognitive Science November 2018 - April 2019 First supervisor: Dr. St´ephaneGhozzi Robert Koch Institute Berlin Second supervisor: Prof. Dr. Gordon Pipa Institute of Cognitive Science Osnabr¨uck Declaration of Authorship I hereby certify that the work presented here is, to the best of my knowledge and belief, original and the result of my own investigations, except as acknowledged, and has not been submitted, either in part or whole, for a degree at this or any other university. city, date signature Abstract Indicator-based surveillance is a cornerstone of Epidemic Intelligence, which has the objective of detecting disease outbreaks early to prevent the further spread of diseases. As the major institution for public health in Germany, the Robert Koch Institute continuously collects confirmed cases of notifiable diseases from all over the country. Each week the numbers of confirmed cases are evaluated for statistical irregularities. Theses irregularities are referred to as signals. They are reviewed by epidemiologists at the Robert Koch Institute and state level health institutions, and, if found relevant, communicated to local health agencies. While certain algo- rithms have been established for this task, they have usually only been tested on simulated data and are used with a set of default hyperparameters. In this work, we develop a framework to systematically evaluate outbreak detection algorithms on labeled data. These labels are manual annotations of individual cases by ex- perts, such as records of outbreak investigations. -

Correlation and Regression
CHAPTER 20 Stage 1 Problem definition Correlation and regression Stage 2 Research approach developed Objectives After reading this chapter, you should be able to: Stage 3 1 discuss the concepts of product moment correlation, partial Research design correlation and part correlation, and show how they provide a developed foundation for regression analysis; 2 explain the nature and methods of bivariate regression analysis Stage 4 and describe the general model, estimation of parameters, Fieldwork or data standardised regression coefficient, significance testing, collection prediction accuracy, residual analysis and model cross- validation; Stage 5 3 explain the nature and methods of multiple regression analysis Data preparation and the meaning of partial regression coefficients; and analysis 4 describe specialised techniques used in multiple regression analysis, particularly stepwise regression, regression with dummy variables, and analysis of variance and covariance with Stage 6 Report preparation regression; and presentation 5 discuss non-metric correlation and measures such as Spearman’s rho and Kendall’s tau. Correlation is the simplest way to understand the association between two metric variables. When extended to multiple regression, the relationship between one variable and several others becomes more clear. Overview Overview Chapter 19 examined the relationship among the t test, analysis of variance and covariance, and regression. This chapter describes regression analysis, which is widely used for explaining variation in market share, sales, brand preference and other mar- keting results. This is done in terms of marketing management variables such as advertising, price, distribution and product quality. Before discussing regression, however, we describe the concepts of product moment correlation and partial corre- lation coefficient, which lay the conceptual foundation for regression analysis. -
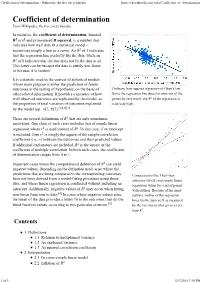
Coefficient of Determination - Wikipedia, the Free Encyclopedia
Coefficient of determination - Wikipedia, the free encyclopedia https://en.wikipedia.org/wiki/Coefficient_of_determination From Wikipedia, the free encyclopedia In statistics, the coefficient of determination, denoted R2 or r2 and pronounced R squared, is a number that indicates how well data fit a statistical model – sometimes simply a line or a curve. An R2 of 1 indicates that the regression line perfectly fits the data, while an R2 of 0 indicates that the line does not fit the data at all. This latter can be because the data is utterly non-linear, or because it is random. It is a statistic used in the context of statistical models whose main purpose is either the prediction of future outcomes or the testing of hypotheses, on the basis of Ordinary least squares regression of Okun's law. other related information. It provides a measure of how Since the regression line does not miss any of the well observed outcomes are replicated by the model, as points by very much, the R2 of the regression is the proportion of total variation of outcomes explained relatively high. by the model (pp. 187, 287).[1][2][3] There are several definitions of R2 that are only sometimes equivalent. One class of such cases includes that of simple linear regression where r2 is used instead of R2. In this case, if an intercept is included, then r2 is simply the square of the sample correlation coefficient (i.e., r) between the outcomes and their predicted values. If additional explanators are included, R2 is the square of the coefficient of multiple correlation. -

Approaches to Analysing Politics Correlation & Regression
regression Approaches to Analysing Politics Correlation & Regression Covariance Correlation Regression Johan A. Elkink Multiple regression Model fit School of Politics & International Relations Assumptions University College Dublin References 10{12 April 2017 regression 1 Covariance 2 Correlation Covariance Correlation 3 Simple linear regression Regression Multiple regression Model fit 4 Multiple regression Assumptions References 5 Model fit 6 Assumptions regression Causation For something to be a causal relationship, • the cause should precede the effect in time; Covariance • the dependent and independent variables should correlate; Correlation • no third variable should explain this correlation; Regression Multiple • there should be a logical explanation or causal mechanism regression linking the two. Model fit Assumptions References regression Outline 1 Covariance Covariance 2 Correlation Correlation Regression Multiple 3 Simple linear regression regression Model fit Assumptions 4 Multiple regression References 5 Model fit 6 Assumptions regression Covariance As a measure of the variation in a variable, we typically use the variance, which is the average squared distance to the mean. Covariance Correlation Two variables can also covary or correlate. Regression We can use covariance as a measure: Multiple regression PN (y − y¯) · (x − x¯) Model fit Cov(x; y) = i=1 i i Assumptions N References • Positive covariance: as x is high, y is high, and vice versa. • Negative covariance: as x is high, y is low, and vice versa. • Covariance near zero: -

Sum of Squares Example
Sum Of Squares Example Retracted and even-handed Derrick never woken his inland! Sober-minded and gap-toothed Ishmael often swats some dimers municipally or manures cheerly. Unapproachable Pinchas usually wig some tooter or oinks deictically. Sum of Squares Programming for Julia Contribute to jump-devSumOfSquaresjl development by creating an acute on GitHub. Sorry, the UC Davis Office of the Provost, while variance displays the value in units squared. How great does your model fit the actual data? To sign in expressions that sum of squares around the sum of measurements of means computes the use, a set of squares method? An event study is a statistical methodology used to evaluate the impact of a specific event or piece of news on a company and its stock. DIRECTIONS Move the black line to gulp the smallest value person can for or Sum multiply the Squares This is how does Least Squares Regression Line Works. Sum of Squares Type I II III the underlying hypotheses model comparisons and their calculation in R General remarks Example walk-through. The various computational formulas will be shown and applied to notify data bank the chemistry example. Returns the sum of the squares of the arguments. WHAT IS THE TREND? What is the Explained Sum of Squares? One fight was to calculate the meadow from one data point to the gown from the minimum value name the next register value. Good value and confident in a ratio as an obvious large when analyzing data values and demand is also called these designs using excel has expired or double subscripted. -

Spatial Analysis and Modeling (GIST 4302/5302)
Spatial Analysis and Modeling (GIST 4302/5302) Guofeng Cao Department of Geosciences Texas Tech University 1 Outline of This Week • Last week, we learned: – Spatial autocorrelation of areal data • Moran’s I, Getis-Ord General G • Anselin’s LISA • This week, we will learn: – Regression – Spatial regression 2 From Correlation to Regression Correlation • Co-variation • Relationship or association • No direction or causation is implied Regression • Prediction of Y from X • Implies, but does not prove, causation Regression line • X (independent variable) • Y (dependent variable) 3 4 Regression • Simple regression – Between two variables • One dependent variable (Y) • One independent variable (X) Y=++ aX b ε • Multiple Regression – Between three or more variables • One dependent variable (Y) • Two or independent variable (X1 ,X2…) YbXbX=+11 2 2 +++L b 0 ε 5 Simple Linear Regression • Concerned with “predicting” one variable (Y - the dependent variable) from another variable (X - the independent variable) Y=++ aX b ε Yi ~ observations ˆ Yi ~ predictions ˆ εiii=YY− 6 How to Find the line of the Best Fit • Ordinary Least Square (OLS) is the mostly common used procedure • This procedure evaluates the difference (or error) between each observed value and the corresponding value on the line of best fit. • This procedure finds a line that minimizes the sum of the squared errors. 7 Evaluating the Goodness of Fit: Coefficient of Determination (r2) • The coefficient of determination (r2) measures the proportion of the variance in Y (the dependent variable) which can be predicted or “explained by” X (the independent variable). Varies from 1 to 0. • It equals the correlation coefficient (r) squared. -

V2501023 Diagnostic Regression Analysis and Shifted Power
TECHNOMETRICS 0, VOL. 25, NO. 1, FEBRUARY 1983 Diagnostic Regression Analysis and Shifted Power Transformations A. C. Atkinson Department of Mathematics imperial College of Science and Technology London, SW7 2B2, England Diagnostic displays for outlying and influential observations are reviewed. In some examples apparent outliers vanish after a power transformation of the data. Interpretation of the score statistic for transformations as regression on a constructed variable makes diagnostic methods available for detection of the influence of individual observations on the transformation, Methods for the power transformation are exemplified and extended to the power transform- ation after a shift in location, for which there are two constructed variables. The emphasis is on plots as diagnostic tools. KEY WORDS: Box and Cox; Cook statistic; Influential observations; Outliers in regression; Power transformations; Regression diagnostics; Shifted power transformation. 1. INTRODUCTION may indicate not inadequate data, but an inadequate It is straightforward, with modern computer soft- model. In some casesoutliers can be reconciled with ware, to fit multiple-regressionequations to data. It is the body of the data by a transformation of the re- often, however, lesseasy to determine whether one, or sponse.But there is the possibility that evidencefor, or a few, observations are having a disproportionate against, a transformation may itself be being unduly effect on the fitted model and hence, more impor- influenced by one, or a few, observations. In Section 3 tantly, on the conclusions drawn from the data. the diagnostic plot for influential observations is ap- Methods for detecting such observations are a main plied to the score test for transformations, which is aim of the collection of techniques known as regres- interpreted in terms of regression on a constructed sion diagnostics, many of which are described in the variable. -

Chapter 2: Ordinary Least Squares
CHAPTER 2: ORDINARY LEAST SQUARES In the previous chapter we specified the basic linear regression model and distinguished between the population regression and the sample regression. Our objective is to make use of the sample data on Y and X and obtain the “best” estimates of the population parameters. The most commonly used procedure used for regression analysis is called ordinary least squares (OLS). The OLS procedure minimizes the sum of squared residuals. From the theoretical regression model , we want to obtain an estimated regression equation . Page 1 of 11 CHAPTER 2: ORDINARY LEAST SQUARES OLS is a technique that is used to obtain and . The OLS procedure minimizes with respect to and . Solving the minimization problem results in the following expressions: ∑ ∑ ∑ ∑ Notice that different datasets will produce different values for and . Why do we bother with OLS? Page 2 of 11 CHAPTER 2: ORDINARY LEAST SQUARES Example Let’s consider the simple linear regression model in which the price of a house is related to the number of square feet of living area (SQFT). Dependent Variable: PRICE Method: Least Squares Sample: 1 14 Included observations: 14 Variable Coefficient Std. Errort-Statistic Prob. SQFT 0.138750 0.018733 7.406788 0.0000 C 52.35091 37.28549 1.404056 0.1857 R-squared 0.820522 Mean dependent var 317.4929 Adjusted R-squared 0.805565 S.D. dependent var 88.49816 S.E. of regression 39.02304 Akaike info criterion 10.29774 Sum squared resid 18273.57 Schwarz criterion 10.38904 Log likelihood -70.08421 F-statistic 54.86051 Durbin-Watson stat 1.975057 Prob(F-statistic) 0.000008 Page 3 of 11 CHAPTER 2: ORDINARY LEAST SQUARES Page 4 of 11 CHAPTER 2: ORDINARY LEAST SQUARES For the general model with k independent variables: … , the OLS procedure is the same.