Ionocytes and CFTR Chloride Channel Expression in Normal and Cystic Fibrosis Nasal and Bronchial Epithelial Cells
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Screening and Identification of Key Biomarkers in Clear Cell Renal Cell Carcinoma Based on Bioinformatics Analysis
bioRxiv preprint doi: https://doi.org/10.1101/2020.12.21.423889; this version posted December 23, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Screening and identification of key biomarkers in clear cell renal cell carcinoma based on bioinformatics analysis Basavaraj Vastrad1, Chanabasayya Vastrad*2 , Iranna Kotturshetti 1. Department of Biochemistry, Basaveshwar College of Pharmacy, Gadag, Karnataka 582103, India. 2. Biostatistics and Bioinformatics, Chanabasava Nilaya, Bharthinagar, Dharwad 580001, Karanataka, India. 3. Department of Ayurveda, Rajiv Gandhi Education Society`s Ayurvedic Medical College, Ron, Karnataka 562209, India. * Chanabasayya Vastrad [email protected] Ph: +919480073398 Chanabasava Nilaya, Bharthinagar, Dharwad 580001 , Karanataka, India bioRxiv preprint doi: https://doi.org/10.1101/2020.12.21.423889; this version posted December 23, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract Clear cell renal cell carcinoma (ccRCC) is one of the most common types of malignancy of the urinary system. The pathogenesis and effective diagnosis of ccRCC have become popular topics for research in the previous decade. In the current study, an integrated bioinformatics analysis was performed to identify core genes associated in ccRCC. An expression dataset (GSE105261) was downloaded from the Gene Expression Omnibus database, and included 26 ccRCC and 9 normal kideny samples. Assessment of the microarray dataset led to the recognition of differentially expressed genes (DEGs), which was subsequently used for pathway and gene ontology (GO) enrichment analysis. -

Dlx3b/4B Is Required for Early-Born but Not Later-Forming Sensory Hair Cells
© 2017. Published by The Company of Biologists Ltd | Biology Open (2017) 6, 1270-1278 doi:10.1242/bio.026211 RESEARCH ARTICLE Dlx3b/4b is required for early-born but not later-forming sensory hair cells during zebrafish inner ear development Simone Schwarzer, Sandra Spieß, Michael Brand and Stefan Hans* ABSTRACT complex arrangement of mechanosensory hair cells, nonsensory Morpholino-mediated knockdown has shown that the homeodomain supporting cells and sensory neurons (Barald and Kelley, 2004; Raft transcription factors Dlx3b and Dlx4b are essential for proper and Groves, 2015; Whitfield, 2015). Inner ear formation is a induction of the otic-epibranchial progenitor domain (OEPD), as multistep process initiated by the establishment of the preplacodal well as subsequent formation of sensory hair cells in the developing region, a zone of ectoderm running around the anterior border of the zebrafish inner ear. However, increasing use of reverse genetic neural plate containing precursors for all sensory placodes (Streit, approaches has revealed poor correlation between morpholino- 2007). The preplacodal region is further specified into a common induced and mutant phenotypes. Using CRISPR/Cas9-mediated otic-epibranchial progenitor domain (OEPD) that in zebrafish also mutagenesis, we generated a defined deletion eliminating the entire contains the progenitors of the anterior lateral line ganglion (Chen open reading frames of dlx3b and dlx4b (dlx3b/4b) and investigated a and Streit, 2013; McCarroll et al., 2012; Hans et al., 2013). potential phenotypic -

Genome-Wide DNA Methylation Analysis of KRAS Mutant Cell Lines Ben Yi Tew1,5, Joel K
www.nature.com/scientificreports OPEN Genome-wide DNA methylation analysis of KRAS mutant cell lines Ben Yi Tew1,5, Joel K. Durand2,5, Kirsten L. Bryant2, Tikvah K. Hayes2, Sen Peng3, Nhan L. Tran4, Gerald C. Gooden1, David N. Buckley1, Channing J. Der2, Albert S. Baldwin2 ✉ & Bodour Salhia1 ✉ Oncogenic RAS mutations are associated with DNA methylation changes that alter gene expression to drive cancer. Recent studies suggest that DNA methylation changes may be stochastic in nature, while other groups propose distinct signaling pathways responsible for aberrant methylation. Better understanding of DNA methylation events associated with oncogenic KRAS expression could enhance therapeutic approaches. Here we analyzed the basal CpG methylation of 11 KRAS-mutant and dependent pancreatic cancer cell lines and observed strikingly similar methylation patterns. KRAS knockdown resulted in unique methylation changes with limited overlap between each cell line. In KRAS-mutant Pa16C pancreatic cancer cells, while KRAS knockdown resulted in over 8,000 diferentially methylated (DM) CpGs, treatment with the ERK1/2-selective inhibitor SCH772984 showed less than 40 DM CpGs, suggesting that ERK is not a broadly active driver of KRAS-associated DNA methylation. KRAS G12V overexpression in an isogenic lung model reveals >50,600 DM CpGs compared to non-transformed controls. In lung and pancreatic cells, gene ontology analyses of DM promoters show an enrichment for genes involved in diferentiation and development. Taken all together, KRAS-mediated DNA methylation are stochastic and independent of canonical downstream efector signaling. These epigenetically altered genes associated with KRAS expression could represent potential therapeutic targets in KRAS-driven cancer. Activating KRAS mutations can be found in nearly 25 percent of all cancers1. -

Supplementary Table 5. Clover Results Indicate the Number Of
Supplementary Table 5. Clover results indicate the number of chromosomes with transcription factor binding motifs statistically over‐ or under‐represented in HTE DHS within intergenic sequence (more than 2kb outside of any gene). Analysis was divided into three groups (all DHS, HTE‐selective DHS, and ubiquitous DHS). Motifs with more than one entry in the databases utilized were edited to retain only the first occurrence of the motif. All DHS x Intergenic TEselective DHS x Intergenic Ubiquitous DHS x Intergenic ID Name p < 0.01 p > 0.99 ID Name p < 0.01 p > 0.99 ID Name p < 0.01 p > 0.99 MA0002.2 RUNX1 23 0 MA0080.2 SPI1 23 0 MA0055.1 Myf 23 0 MA0003.1 TFAP2A 23 0 MA0089.1 NFE2L1::MafG 23 0 MA0068.1 Pax4 23 0 MA0039.2 Klf4 23 0 MA0098.1 ETS1 23 0 MA0080.2 SPI1 23 0 MA0055.1 Myf 23 0 MA0099.2 AP1 23 0 MA0098.1 ETS1 23 0 MA0056.1 MZF1_1‐4 23 0 MA0136.1 ELF5 23 0 MA0139.1 CTCF 23 0 MA0079.2 SP1 23 0 MA0145.1 Tcfcp2l1 23 0 V$ALX3_01 ALX‐3 23 0 MA0080.2 SPI1 23 0 MA0150.1 NFE2L2 23 0 V$ALX4_02 Alx‐4 23 0 myocyte enhancer MA0081.1 SPIB 23 0 MA0156.1 FEV 23 0 V$AMEF2_Q6 factor 23 0 MA0089.1 NFE2L1::MafG 23 0 V$AP1FJ_Q2 activator protein 1 23 0 V$AP1_01 AP‐1 23 0 MA0090.1 TEAD1 23 0 V$AP4_Q5 activator protein 4 23 0 V$AP2_Q6_01 AP‐2 23 0 MA0098.1 ETS1 23 0 V$AR_Q6 half‐site matrix 23 0 V$ARX_01 Arx 23 0 BTB and CNC homolog MA0099.2 AP1 23 0 V$BACH1_01 1 23 0 V$BARHL1_01 Barhl‐1 23 0 BTB and CNC homolog MA0136.1 ELF5 23 0 V$BACH2_01 2 23 0 V$BARHL2_01 Barhl2 23 0 MA0139.1 CTCF 23 0 V$CMAF_02 C‐MAF 23 0 V$BARX1_01 Barx1 23 0 MA0144.1 Stat3 23 0 -
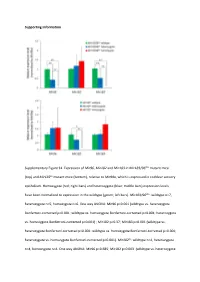
Supporting Information Supplementary Figure S1
Supporting Information Supplementary Figure S1. Expression of Mir96, Mir182 and Mir183 in Mir183/96dko mutant mice (top) and Mir182ko mutant mice (bottom), relative to Mir99a, which is expressed in cochlear sensory epithelium. Homozygote (red; right bars) and heterozygote (blue; middle bars) expression levels have been normalised to expression in the wildtype (green; left bars). Mir183/96dko: wildtype n=7, heterozygote n=5, homozygote n=6. One way ANOVA: Mir96 p<0.001 (wildtype vs. heterozygote Bonferroni‐corrected p<0.001; wildtype vs. homozygote Bonferroni‐corrected p<0.001; heterozygote vs. homozygote Bonferroni‐corrected p=0.001) ; Mir182 p=0.37; Mir183 p<0.001 (wildtype vs. heterozygote Bonferroni‐corrected p=0.001; wildtype vs. homozygote Bonferroni‐corrected p<0.001; heterozygote vs. homozygote Bonferroni‐corrected p<0.001). Mir182ko: wildtype n=4, heterozygote n=4, homozygote n=4. One way ANOVA: Mir96 p=0.685; Mir182 p=0.003 (wildtype vs. heterozygote Bonferroni‐corrected p=0.397; wildtype vs. homozygote Bonferroni‐corrected p=0.003; heterozygote vs. homozygote Bonferroni‐corrected p=0.032); Mir183 p=0.04 (wildtype vs. heterozygote Bonferroni‐corrected p=1.0; wildtype vs. homozygote Bonferroni‐corrected p=0.068; heterozygote vs. homozygote Bonferroni‐corrected p=0.094), Error bars are standard deviation (* = P < 0.05, ** = P ≤ 0.01). Supplementary Figure S2. Individual ABR thresholds of wildtype, heterozygous and homozygous Mir183/96dko mice at all ages tested. Number of mice of each genotype tested at each age is shown on the threshold plot. Supplementary Figure S3. Individual ABR thresholds of wildtype, heterozygous and homozygous Mir182ko mice at all ages tested. -

Supplement. Transcriptional Factors (TF), Protein Name and Their Description Or Function
Supplement. Transcriptional factors (TF), protein name and their description or function. TF Protein name TF description/function ARID3A AT rich interactive domain 3A (BRIGHT-like) This gene encodes a member of the ARID (AT-rich interaction domain) family of DNA binding proteins. ATF4 Activating Transcription Factor 4 Transcriptional activator. Binds the cAMP response element (CRE) (consensus: 5-GTGACGT[AC][AG]-3), a sequence present in many viral and cellular promoters. CTCF CCCTC-Binding Factor Chromatin binding factor that binds to DNA sequence specific sites. Involved in transcriptional regulation by binding to chromatin insulators and preventing interaction between promoter and nearby enhancers and silencers. The protein can bind a histone acetyltransferase (HAT)-containing complex and function as a transcriptional activator or bind a histone deacetylase (HDAC)-containing complex and function as a transcriptional repressor. E2F1-6 E2F transcription factors 1-6 The protein encoded by this gene is a member of the E2F family of transcription factors. The E2F family plays a crucial role in the control of cell cycle and action of tumor suppressor proteins and is also a target of the transforming proteins of small DNA tumor viruses. The E2F proteins contain several evolutionally conserved domains found in most members of the family. These domains include a DNA binding domain, a dimerization domain which determines interaction with the differentiation regulated transcription factor proteins (DP), a transactivation domain enriched in acidic amino acids, and a tumor suppressor protein association domain which is embedded within the transactivation domain. EBF1 Transcription factor COE1 EBF1 has been shown to interact with ZNF423 and CREB binding proteins. -

Engineered Type 1 Regulatory T Cells Designed for Clinical Use Kill Primary
ARTICLE Acute Myeloid Leukemia Engineered type 1 regulatory T cells designed Ferrata Storti Foundation for clinical use kill primary pediatric acute myeloid leukemia cells Brandon Cieniewicz,1* Molly Javier Uyeda,1,2* Ping (Pauline) Chen,1 Ece Canan Sayitoglu,1 Jeffrey Mao-Hwa Liu,1 Grazia Andolfi,3 Katharine Greenthal,1 Alice Bertaina,1,4 Silvia Gregori,3 Rosa Bacchetta,1,4 Norman James Lacayo,1 Alma-Martina Cepika1,4# and Maria Grazia Roncarolo1,2,4# Haematologica 2021 Volume 106(10):2588-2597 1Department of Pediatrics, Division of Stem Cell Transplantation and Regenerative Medicine, Stanford School of Medicine, Stanford, CA, USA; 2Stanford Institute for Stem Cell Biology and Regenerative Medicine, Stanford School of Medicine, Stanford, CA, USA; 3San Raffaele Telethon Institute for Gene Therapy, Milan, Italy and 4Center for Definitive and Curative Medicine, Stanford School of Medicine, Stanford, CA, USA *BC and MJU contributed equally as co-first authors #AMC and MGR contributed equally as co-senior authors ABSTRACT ype 1 regulatory (Tr1) T cells induced by enforced expression of interleukin-10 (LV-10) are being developed as a novel treatment for Tchemotherapy-resistant myeloid leukemias. In vivo, LV-10 cells do not cause graft-versus-host disease while mediating graft-versus-leukemia effect against adult acute myeloid leukemia (AML). Since pediatric AML (pAML) and adult AML are different on a genetic and epigenetic level, we investigate herein whether LV-10 cells also efficiently kill pAML cells. We show that the majority of primary pAML are killed by LV-10 cells, with different levels of sensitivity to killing. Transcriptionally, pAML sensitive to LV-10 killing expressed a myeloid maturation signature. -

Transcriptomic Profiling Identifies Differentially Expressed Genes In
G C A T T A C G G C A T genes Article Transcriptomic Profiling Identifies Differentially Expressed Genes in Palbociclib-Resistant ER+ MCF7 Breast Cancer Cells Lilibeth Lanceta 1, Conor O’Neill 2, Nadiia Lypova 1 , Xiahong Li 3, Eric Rouchka 4, Sabine Waigel 1, Jorge G. Gomez-Gutierrez 5,6 , Jason Chesney 1,6 and Yoannis Imbert-Fernandez 1,6,* 1 Department of Medicine, School of Medicine, University of Louisville, Louisville, KY 40202, USA; [email protected] (L.L.); [email protected] (N.L.); [email protected] (S.W.); [email protected] (J.C.) 2 College of Medicine, University of Kentucky, Lexington, KY 40506, USA; [email protected] 3 Department of Anatomical Sciences and Neurobiology, Bioinformatics Core, University of Louisville, Louisville, KY 40202, USA; [email protected] 4 Department of Computer Engineering and Computer Science, University of Louisville, Louisville, KY 40292, USA; [email protected] 5 Department of Surgery, School of Medicine, University of Louisville, Louisville, KY 40202, USA; [email protected] 6 James Graham Brown Cancer Center School of Medicine, University of Louisville, Louisville, KY 40202, USA * Correspondence: [email protected]; Tel.: +1-502-852-6570 Received: 20 March 2020; Accepted: 18 April 2020; Published: 24 April 2020 Abstract: Acquired resistance to cyclin-dependent kinases 4 and 6 (CDK4/6) inhibition in estrogen receptor-positive (ER+) breast cancer remains a significant clinical challenge. Efforts to uncover the mechanisms underlying resistance are needed to establish clinically actionable targets effective against resistant tumors. In this study, we sought to identify differentially expressed genes (DEGs) associated with acquired resistance to palbociclib in ER+ breast cancer. -

Perkinelmer Genomics to Request the Saliva Swab Collection Kit for Patients That Cannot Provide a Blood Sample As Whole Blood Is the Preferred Sample
Autism and Intellectual Disability TRIO Panel Test Code TR002 Test Summary This test analyzes 2429 genes that have been associated with Autism and Intellectual Disability and/or disorders associated with Autism and Intellectual Disability with the analysis being performed as a TRIO Turn-Around-Time (TAT)* 3 - 5 weeks Acceptable Sample Types Whole Blood (EDTA) (Preferred sample type) DNA, Isolated Dried Blood Spots Saliva Acceptable Billing Types Self (patient) Payment Institutional Billing Commercial Insurance Indications for Testing Comprehensive test for patients with intellectual disability or global developmental delays (Moeschler et al 2014 PMID: 25157020). Comprehensive test for individuals with multiple congenital anomalies (Miller et al. 2010 PMID 20466091). Patients with autism/autism spectrum disorders (ASDs). Suspected autosomal recessive condition due to close familial relations Previously negative karyotyping and/or chromosomal microarray results. Test Description This panel analyzes 2429 genes that have been associated with Autism and ID and/or disorders associated with Autism and ID. Both sequencing and deletion/duplication (CNV) analysis will be performed on the coding regions of all genes included (unless otherwise marked). All analysis is performed utilizing Next Generation Sequencing (NGS) technology. CNV analysis is designed to detect the majority of deletions and duplications of three exons or greater in size. Smaller CNV events may also be detected and reported, but additional follow-up testing is recommended if a smaller CNV is suspected. All variants are classified according to ACMG guidelines. Condition Description Autism Spectrum Disorder (ASD) refers to a group of developmental disabilities that are typically associated with challenges of varying severity in the areas of social interaction, communication, and repetitive/restricted behaviors. -

Table S4 Downregulated Transcription Factors That Recovered by 3 Hours
Table S4 Downregulated transcription factors that recovered by 3 hours post-treatment log2FC log2FC Log2FC Pubmed NCBI Short DNA Binding Trinity ID AEP ID Gene Model NCBI Description 0h 3h 6h Hit Accession Name Domain Hydra vulgaris MicrophthalmIa- TRINITY_DN7608_c0_g1 t22195aep Sc4wPfr_552.g10536.t1 -0.326 0 0 NA CDG71838.1 assocIated TFE3 bHLH transcrIption factor [Hydra vulgarIs] PREDICTED: zInc finger proteIn 37-like TRINITY_DN10125_c0_g1 t26873aep Sc4wPfr_1909.g11470.t1 -0.641 0 0 NA XP_012558352.1 ZSC31 zf-C2H2 isoform X1 [Hydra vulgarIs] PREDICTED: transcrIption factor TRINITY_DN14709_c0_g1 t5528aep Sc4wPfr_59.2.g12567.t1 -0.925 0 0 NA XP_002154370.1 HySox19a HMG box Sox-19a-like [Hydra vulgarIs] PREDICTED: uncharacterIzed proteIn TRINITY_DN5359_c0_g1 t23172aep Sc4wPfr_297.g13156.t1 -1.507 0 0 NA XP_012555836.1 LOC101236863 Sox14 HMG box [Hydra vulgarIs], simIlar to SoxB3 Hydractinia echinata Hydra vulgarIs ZInc finger proteIn ZIC 5, TRINITY_DN4294_c0_g1 t20709aep Sc4wPfr_237.2.g16165.t1 -1.072 0 0 NA CDG67849.1 ZIc5-like zf-C2H2 partIal [Hydra vulgarIs] Hydra vulgaris Putative TRINITY_DN37877_c0_g1 t26616aep Sc4wPfr_14.g1768.t2 -1.427 0 0 NA CDG72115.1 transcrIption factor NA zf-C2H2 Ovo-like 1, partial [Hydra vulgarIs] CnAlx arIstaless-like protein TRINITY_DN5649_c0_g1 t16456aep Sc4wPfr_654.g18227.t1 -1.160 0 0 NA AAG03082.1 (SmIth, Homeobox [Hydra vulgarIs] 2000) PREDICTED: uncharacterIzed TRINITY_DN3178_c1_g1 t17964aep Sc4wPfr_68.g2328.t1 -1.605 0 0 NA XP_012567188.1 proteIn Jun bZIP LOC105851038 [Hydra vulgarIs] Hydra vulgaris -

Molecular Targeting and Enhancing Anticancer Efficacy of Oncolytic HSV-1 to Midkine Expressing Tumors
University of Cincinnati Date: 12/20/2010 I, Arturo R Maldonado , hereby submit this original work as part of the requirements for the degree of Doctor of Philosophy in Developmental Biology. It is entitled: Molecular Targeting and Enhancing Anticancer Efficacy of Oncolytic HSV-1 to Midkine Expressing Tumors Student's name: Arturo R Maldonado This work and its defense approved by: Committee chair: Jeffrey Whitsett Committee member: Timothy Crombleholme, MD Committee member: Dan Wiginton, PhD Committee member: Rhonda Cardin, PhD Committee member: Tim Cripe 1297 Last Printed:1/11/2011 Document Of Defense Form Molecular Targeting and Enhancing Anticancer Efficacy of Oncolytic HSV-1 to Midkine Expressing Tumors A dissertation submitted to the Graduate School of the University of Cincinnati College of Medicine in partial fulfillment of the requirements for the degree of DOCTORATE OF PHILOSOPHY (PH.D.) in the Division of Molecular & Developmental Biology 2010 By Arturo Rafael Maldonado B.A., University of Miami, Coral Gables, Florida June 1993 M.D., New Jersey Medical School, Newark, New Jersey June 1999 Committee Chair: Jeffrey A. Whitsett, M.D. Advisor: Timothy M. Crombleholme, M.D. Timothy P. Cripe, M.D. Ph.D. Dan Wiginton, Ph.D. Rhonda D. Cardin, Ph.D. ABSTRACT Since 1999, cancer has surpassed heart disease as the number one cause of death in the US for people under the age of 85. Malignant Peripheral Nerve Sheath Tumor (MPNST), a common malignancy in patients with Neurofibromatosis, and colorectal cancer are midkine- producing tumors with high mortality rates. In vitro and preclinical xenograft models of MPNST were utilized in this dissertation to study the role of midkine (MDK), a tumor-specific gene over- expressed in these tumors and to test the efficacy of a MDK-transcriptionally targeted oncolytic HSV-1 (oHSV). -

(12) Patent Application Publication (10) Pub. No.: US 2009/0269772 A1 Califano Et Al
US 20090269772A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2009/0269772 A1 Califano et al. (43) Pub. Date: Oct. 29, 2009 (54) SYSTEMS AND METHODS FOR Publication Classification IDENTIFYING COMBINATIONS OF (51) Int. Cl. COMPOUNDS OF THERAPEUTIC INTEREST CI2O I/68 (2006.01) CI2O 1/02 (2006.01) (76) Inventors: Andrea Califano, New York, NY G06N 5/02 (2006.01) (US); Riccardo Dalla-Favera, New (52) U.S. Cl. ........... 435/6: 435/29: 706/54; 707/E17.014 York, NY (US); Owen A. (57) ABSTRACT O'Connor, New York, NY (US) Systems, methods, and apparatus for searching for a combi nation of compounds of therapeutic interest are provided. Correspondence Address: Cell-based assays are performed, each cell-based assay JONES DAY exposing a different sample of cells to a different compound 222 EAST 41ST ST in a plurality of compounds. From the cell-based assays, a NEW YORK, NY 10017 (US) Subset of the tested compounds is selected. For each respec tive compound in the Subset, a molecular abundance profile from cells exposed to the respective compound is measured. (21) Appl. No.: 12/432,579 Targets of transcription factors and post-translational modu lators of transcription factor activity are inferred from the (22) Filed: Apr. 29, 2009 molecular abundance profile data using information theoretic measures. This data is used to construct an interaction net Related U.S. Application Data work. Variances in edges in the interaction network are used to determine the drug activity profile of compounds in the (60) Provisional application No. 61/048.875, filed on Apr.