Paleoceanographic Changes in the Late Pliocene Promoted Rapid
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
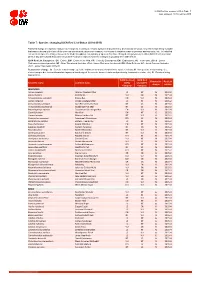
Table 7: Species Changing IUCN Red List Status (2014-2015)
IUCN Red List version 2015.4: Table 7 Last Updated: 19 November 2015 Table 7: Species changing IUCN Red List Status (2014-2015) Published listings of a species' status may change for a variety of reasons (genuine improvement or deterioration in status; new information being available that was not known at the time of the previous assessment; taxonomic changes; corrections to mistakes made in previous assessments, etc. To help Red List users interpret the changes between the Red List updates, a summary of species that have changed category between 2014 (IUCN Red List version 2014.3) and 2015 (IUCN Red List version 2015-4) and the reasons for these changes is provided in the table below. IUCN Red List Categories: EX - Extinct, EW - Extinct in the Wild, CR - Critically Endangered, EN - Endangered, VU - Vulnerable, LR/cd - Lower Risk/conservation dependent, NT - Near Threatened (includes LR/nt - Lower Risk/near threatened), DD - Data Deficient, LC - Least Concern (includes LR/lc - Lower Risk, least concern). Reasons for change: G - Genuine status change (genuine improvement or deterioration in the species' status); N - Non-genuine status change (i.e., status changes due to new information, improved knowledge of the criteria, incorrect data used previously, taxonomic revision, etc.); E - Previous listing was an Error. IUCN Red List IUCN Red Reason for Red List Scientific name Common name (2014) List (2015) change version Category Category MAMMALS Aonyx capensis African Clawless Otter LC NT N 2015-2 Ailurus fulgens Red Panda VU EN N 2015-4 -

Status and Conservation of Shearwaters of the North Pacific
Status and conservation of shearwaters Reprinted from Vermeer, K.; Briggs, K.T.; Morgan, K.H.; Siegel-Causey, D. (eds.). 1993. The status, ecology. and of the North Pacific conservation of marine birds of the North Pacific. Can. Wildl. Sen. Spec. Publ., Ottawa. William T. Everett’ and Robert L. Pitman’ ‘WesreniFoundarion of Vertebrate Zoolog).. 1100 Glendon Avenue, Los Angeies, CA 90024 ’Sowhwesr Fisheries Science Center,P.O. Box 271, La Jolla, CA 92038 Abstract level of effort expended on pelagic studies of many seabirds, including shearwaters, has increased dramatically, yielding Of the shearwaters that breed in the North Pacific, many new facts on behaviour, habitat selection, migratory the species in the most peril at present is Townsend’s routes and timing, feeding habits, and mortality. Shearwater Pufinus ourtcuforis. Feral cats and pigs pose a In this paper we discuss the general biology and status threat on its breeding grounds. More information is needed of each of the shearwater species occurring in the North Pacific, on its current status, and that of the Black-vented Shearwater consider threats and hazards both on land and at sea, and offer P. opisthomefas,most of whose nesting colonies have well- recommendations for future study and conservation efforts. established feral cat populations. Efforts are being made to protect Newell’s Shearwater P. newelli in the Hawaiian Islands. 2. Species accounts The single greatest threat to migrant shearwaters in the North Pacific is drift gillnet fisheries, which kill hundreds of 2.1. Calonectris leuromelas Streaked Shearwater thousands of birds annually. This species breeds in the far-western Pacific at many locations from the Ryukyu and Bonin islands north to Qingdao RhmC Island in the Yellow Sea, islands off northern Honshu, Japan (Melville 1984; Hasegawa 1984), and has nested on Kmamizin De tous les puffins qui se reproduisent dans le Pacifique Island in the former U.S.S.R.(Litvinenko 1976). -

Tinamiformes – Falconiformes
LIST OF THE 2,008 BIRD SPECIES (WITH SCIENTIFIC AND ENGLISH NAMES) KNOWN FROM THE A.O.U. CHECK-LIST AREA. Notes: "(A)" = accidental/casualin A.O.U. area; "(H)" -- recordedin A.O.U. area only from Hawaii; "(I)" = introducedinto A.O.U. area; "(N)" = has not bred in A.O.U. area but occursregularly as nonbreedingvisitor; "?" precedingname = extinct. TINAMIFORMES TINAMIDAE Tinamus major Great Tinamou. Nothocercusbonapartei Highland Tinamou. Crypturellus soui Little Tinamou. Crypturelluscinnamomeus Thicket Tinamou. Crypturellusboucardi Slaty-breastedTinamou. Crypturellus kerriae Choco Tinamou. GAVIIFORMES GAVIIDAE Gavia stellata Red-throated Loon. Gavia arctica Arctic Loon. Gavia pacifica Pacific Loon. Gavia immer Common Loon. Gavia adamsii Yellow-billed Loon. PODICIPEDIFORMES PODICIPEDIDAE Tachybaptusdominicus Least Grebe. Podilymbuspodiceps Pied-billed Grebe. ?Podilymbusgigas Atitlan Grebe. Podicepsauritus Horned Grebe. Podicepsgrisegena Red-neckedGrebe. Podicepsnigricollis Eared Grebe. Aechmophorusoccidentalis Western Grebe. Aechmophorusclarkii Clark's Grebe. PROCELLARIIFORMES DIOMEDEIDAE Thalassarchechlororhynchos Yellow-nosed Albatross. (A) Thalassarchecauta Shy Albatross.(A) Thalassarchemelanophris Black-browed Albatross. (A) Phoebetriapalpebrata Light-mantled Albatross. (A) Diomedea exulans WanderingAlbatross. (A) Phoebastriaimmutabilis Laysan Albatross. Phoebastrianigripes Black-lootedAlbatross. Phoebastriaalbatrus Short-tailedAlbatross. (N) PROCELLARIIDAE Fulmarus glacialis Northern Fulmar. Pterodroma neglecta KermadecPetrel. (A) Pterodroma -

Mexico Chiapas 15Th April to 27Th April 2021 (13 Days)
Mexico Chiapas 15th April to 27th April 2021 (13 days) Horned Guan by Adam Riley Chiapas is the southernmost state of Mexico, located on the border of Guatemala. Our 13 day tour of Chiapas takes in the very best of the areas birding sites such as San Cristobal de las Casas, Comitan, the Sumidero Canyon, Isthmus of Tehuantepec, Tapachula and Volcan Tacana. A myriad of beautiful and sought after species includes the amazing Giant Wren, localized Nava’s Wren, dainty Pink-headed Warbler, Rufous-collared Thrush, Garnet-throated and Amethyst-throated Hummingbird, Rufous-browed Wren, Blue-and-white Mockingbird, Bearded Screech Owl, Slender Sheartail, Belted Flycatcher, Red-breasted Chat, Bar-winged Oriole, Lesser Ground Cuckoo, Lesser Roadrunner, Cabanis’s Wren, Mayan Antthrush, Orange-breasted and Rose-bellied Bunting, West Mexican Chachalaca, Citreoline Trogon, Yellow-eyed Junco, Unspotted Saw-whet Owl and Long- tailed Sabrewing. Without doubt, the tour highlight is liable to be the incredible Horned Guan. While searching for this incomparable species, we can expect to come across a host of other highlights such as Emerald-chinned, Wine-throated and Azure-crowned Hummingbird, Cabanis’s Tanager and at night the haunting Fulvous Owl! RBL Mexico – Chiapas Itinerary 2 THE TOUR AT A GLANCE… THE ITINERARY Day 1 Arrival in Tuxtla Gutierrez, transfer to San Cristobal del las Casas Day 2 San Cristobal to Comitan Day 3 Comitan to Tuxtla Gutierrez Days 4, 5 & 6 Sumidero Canyon and Eastern Sierra tropical forests Day 7 Arriaga to Mapastepec via the Isthmus of Tehuantepec Day 8 Mapastepec to Tapachula Day 9 Benito Juarez el Plan to Chiquihuites Day 10 Chiquihuites to Volcan Tacana high camp & Horned Guan Day 11 Volcan Tacana high camp to Union Juarez Day 12 Union Juarez to Tapachula Day 13 Final departures from Tapachula TOUR MAP… RBL Mexico – Chiapas Itinerary 3 THE TOUR IN DETAIL… Day 1: Arrival in Tuxtla Gutierrez, transfer to San Cristobal del las Casas. -

Listing Five Foreign Bird Species in Colombia and Ecuador, South America, As Endangered Throughout Their Range; Final Rule
Vol. 78 Tuesday, No. 209 October 29, 2013 Part IV Department of the Interior Fish and Wildlife Service 50 CFR Part 17 Endangered and Threatened Wildlife and Plants; Listing Five Foreign Bird Species in Colombia and Ecuador, South America, as Endangered Throughout Their Range; Final Rule VerDate Mar<15>2010 18:44 Oct 28, 2013 Jkt 232001 PO 00000 Frm 00001 Fmt 4717 Sfmt 4717 E:\FR\FM\29OCR4.SGM 29OCR4 mstockstill on DSK4VPTVN1PROD with RULES4 64692 Federal Register / Vol. 78, No. 209 / Tuesday, October 29, 2013 / Rules and Regulations DEPARTMENT OF THE INTERIOR endangered or threatened we are proposed for these five foreign bird required to publish in the Federal species as endangered, following careful Fish and Wildlife Service Register a proposed rule to list the consideration of all comments we species and, within 1 year of received during the public comment 50 CFR Part 17 publication of the proposed rule, a final periods. rule to add the species to the Lists of [Docket No. FWS–R9–IA–2009–12; III. Costs and Benefits 4500030115] Endangered and Threatened Wildlife and Plants. On July 7, 2009, we We have not analyzed the costs or RIN 1018–AV75 published a proposed rule in which we benefits of this rulemaking action determined that the blue-billed because the Act precludes consideration Endangered and Threatened Wildlife curassow, brown-banded antpitta, Cauca of such impacts on listing and delisting and Plants; Listing Five Foreign Bird guan, gorgeted wood-quail, and determinations. Instead, listing and Species in Colombia and Ecuador, Esmeraldas woodstar currently face delisting decisions are based solely on South America, as Endangered numerous threats and warrant listing the best scientific and commercial Throughout Their Range under the Act as endangered species (74 information available regarding the AGENCY: Fish and Wildlife Service, FR 32308). -

RESTRICTED ANIMAL LIST (Part A) §4-71-6.5 SCIENTIFIC NAME
RESTRICTED ANIMAL LIST (Part A) §4-71-6.5 SCIENTIFIC NAME COMMON NAME §4-71-6.5 LIST OF RESTRICTED ANIMALS September 25, 2018 PART A: FOR RESEARCH AND EXHIBITION SCIENTIFIC NAME COMMON NAME INVERTEBRATES PHYLUM Annelida CLASS Hirudinea ORDER Gnathobdellida FAMILY Hirudinidae Hirudo medicinalis leech, medicinal ORDER Rhynchobdellae FAMILY Glossiphoniidae Helobdella triserialis leech, small snail CLASS Oligochaeta ORDER Haplotaxida FAMILY Euchytraeidae Enchytraeidae (all species in worm, white family) FAMILY Eudrilidae Helodrilus foetidus earthworm FAMILY Lumbricidae Lumbricus terrestris earthworm Allophora (all species in genus) earthworm CLASS Polychaeta ORDER Phyllodocida 1 RESTRICTED ANIMAL LIST (Part A) §4-71-6.5 SCIENTIFIC NAME COMMON NAME FAMILY Nereidae Nereis japonica lugworm PHYLUM Arthropoda CLASS Arachnida ORDER Acari FAMILY Phytoseiidae Iphiseius degenerans predator, spider mite Mesoseiulus longipes predator, spider mite Mesoseiulus macropilis predator, spider mite Neoseiulus californicus predator, spider mite Neoseiulus longispinosus predator, spider mite Typhlodromus occidentalis mite, western predatory FAMILY Tetranychidae Tetranychus lintearius biocontrol agent, gorse CLASS Crustacea ORDER Amphipoda FAMILY Hyalidae Parhyale hawaiensis amphipod, marine ORDER Anomura FAMILY Porcellanidae Petrolisthes cabrolloi crab, porcelain Petrolisthes cinctipes crab, porcelain Petrolisthes elongatus crab, porcelain Petrolisthes eriomerus crab, porcelain Petrolisthes gracilis crab, porcelain Petrolisthes granulosus crab, porcelain Petrolisthes -

Alpha Codes for 2168 Bird Species (And 113 Non-Species Taxa) in Accordance with the 62Nd AOU Supplement (2021), Sorted Taxonomically
Four-letter (English Name) and Six-letter (Scientific Name) Alpha Codes for 2168 Bird Species (and 113 Non-Species Taxa) in accordance with the 62nd AOU Supplement (2021), sorted taxonomically Prepared by Peter Pyle and David F. DeSante The Institute for Bird Populations www.birdpop.org ENGLISH NAME 4-LETTER CODE SCIENTIFIC NAME 6-LETTER CODE Highland Tinamou HITI Nothocercus bonapartei NOTBON Great Tinamou GRTI Tinamus major TINMAJ Little Tinamou LITI Crypturellus soui CRYSOU Thicket Tinamou THTI Crypturellus cinnamomeus CRYCIN Slaty-breasted Tinamou SBTI Crypturellus boucardi CRYBOU Choco Tinamou CHTI Crypturellus kerriae CRYKER White-faced Whistling-Duck WFWD Dendrocygna viduata DENVID Black-bellied Whistling-Duck BBWD Dendrocygna autumnalis DENAUT West Indian Whistling-Duck WIWD Dendrocygna arborea DENARB Fulvous Whistling-Duck FUWD Dendrocygna bicolor DENBIC Emperor Goose EMGO Anser canagicus ANSCAN Snow Goose SNGO Anser caerulescens ANSCAE + Lesser Snow Goose White-morph LSGW Anser caerulescens caerulescens ANSCCA + Lesser Snow Goose Intermediate-morph LSGI Anser caerulescens caerulescens ANSCCA + Lesser Snow Goose Blue-morph LSGB Anser caerulescens caerulescens ANSCCA + Greater Snow Goose White-morph GSGW Anser caerulescens atlantica ANSCAT + Greater Snow Goose Intermediate-morph GSGI Anser caerulescens atlantica ANSCAT + Greater Snow Goose Blue-morph GSGB Anser caerulescens atlantica ANSCAT + Snow X Ross's Goose Hybrid SRGH Anser caerulescens x rossii ANSCAR + Snow/Ross's Goose SRGO Anser caerulescens/rossii ANSCRO Ross's Goose -

SWFSC Archive
CONTENTS LIST OF TABLES ........................................................................................................................ ii LIST OF FIGURES ...................................................................................................................... ii INTRODUCTION......................................................................................................................... 1 OBJECTIVES ............................................................................................................................... 1 STUDY AREA AND ITINERARY ............................................................................................. 2 MATERIALS AND METHODS ................................................................................................. 3 Oceanography ............................................................................................................................. 3 Net Tows ...................................................................................................................................... 4 Acoustic Backscatter.................................................................................................................... 4 Seabirds ....................................................................................................................................... 5 Contour Plots............................................................................................................................... 5 RESULTS ..................................................................................................................................... -

A Sight Record of a Streaked Shearwater in Oregon
NOTES A SIGHT RECORD OF A STREAKED SHEARWATER IN OREGON MICHAEL P. FORCE, 2304 PrinceAlbert Street,Vancouver, B.C., CanadaV5T 3W5 RICHARD A. ROWLETT, P.O. Box 7386, Bellevue,Washington, USA 98008-1386 GEOFF GRACE, 3436 CanberraStreet, SilverSpring, Maryland,USA 20904 On 13 September 1996, while conductingsurveys for marine mammalsand seabirdsaboard the NOAA ship McArthur about 57 kilometersoff the southern Oregon coast,we founda StreakedShearwater (Calonectris leucomelas), a species familiar to both Force and Rowlett. The bird was seen at 08:40 in a large mixed feedingflock over Heceta Bank. Lane County,Oregon (43 ø 59.1 • N, 124 ø 51.4 fW). This constitutesthe firstrecord of the StreakedShearwater for Oregonand the most northeasterlyPacific Ocean occurrence. We hadunobstructed views from the ship'sflying bridge, 14 metersabove sea level. The birdwas seen clearly for abouttwo minutesand passed in frontof the shipas close as 250 meters.It patrolledlow over the water on the starboardside, passed in front of the ship, and eventuallydisappeared astern. We had a variety of binoculars available:20 x 60 prism-stabilizedZeiss (Force), 25 x 150 ship-mountedFujinons (Rowlett),and 7 x 50 hand-heldFujinons (Grace). The ship was engagedin a line- transectsurvey of marine mammalswhose protocol prevented further investigation. We preparedfield notes immediatelyafter the bird was lost from view and before consultingany references.A written report is on file with the Oregon Bird Records Committee. General Appearance:The bird appearedto be in freshplumage with no signof molt, suggestingthat it may have beenin its firstyear. Fairly large and long-winged, gleamingwhite below and brown abovewith a strikingwhite head when seenat a distance.Pale-tipped back feathersgave it a saddledappearance. -

Recovery of the Trinidad Piping-Guan ( Pipile Pipile )
Expedition ID: 300605 Grant Category : Gold Grant Amount: $14,500 Project Pawi: Recovery of the Trinidad Piping-Guan ( Pipile pipile ) Team members: Jahson Alemu, Emma Davis, Aidan Keane, Kevin Mahabir, Srishti Mohais, Alesha Naranjit, Kerrie Naranjit, Bethany Stoker Contact: Aidan Keane Imperial College London Silwood Park Buckhurst Road Ascot Berks SL5 7PY [email protected] Acknowledgements The team would like to thank British Petroleum (BP), BirdLife International, Fauna and Flora International (FFI), Conservation International, Wildlife Conservation Society for funding the project. Special thanks go to Phil McGowan at the World Pheasant Association and to Marianne, Robyn and Kate at the BP Conservation Programme whose help and patience have been invaluable throughout. Project Pawi was only possible because of the generosity, support and encouragement of a large number of people within Trinidad itself, to whom we are very grateful. The team would particularly like to thank the members of the Pawi Study Group, tirelessly led by John and Margaret Cooper, for their practical help, thoughtful discussions and friendship. On ecological matters the guidance and expertise of Adrian Hailey and Howard Nelson was greatly appreciated. Grande Riviere has been at the heart of efforts to study the Pawi for many years, and once again played a central role in our project. We are indebted to owners of the Grande Riviere house for their generosity in allowing us to use the site as our base. The team would especially like to thank the guides of the Grande Riviere Nature Tour Guide Association and Mr Guy for their help and friendship. We are very grateful for the practical support provided by the University of the West Indies, Asa Wright, the Forestry Department, the Wildlife Section and the National Parks Section. -

Conservation Status of New Zealand Birds, 2008
Notornis, 2008, Vol. 55: 117-135 117 0029-4470 © The Ornithological Society of New Zealand, Inc. Conservation status of New Zealand birds, 2008 Colin M. Miskelly* Wellington Conservancy, Department of Conservation, P.O. Box 5086, Wellington 6145, New Zealand [email protected] JOHN E. DOWDING DM Consultants, P.O. Box 36274, Merivale, Christchurch 8146, New Zealand GRAEME P. ELLIOTT Research & Development Group, Department of Conservation, Private Bag 5, Nelson 7042, New Zealand RODNEY A. HITCHMOUGH RALPH G. POWLESLAND HUGH A. ROBERTSON Research & Development Group, Department of Conservation, P.O. Box 10420, Wellington 6143, New Zealand PAUL M. SAGAR National Institute of Water & Atmospheric Research, P.O. Box 8602, Christchurch 8440, New Zealand R. PAUL SCOFIELD Canterbury Museum, Rolleston Ave, Christchurch 8001, New Zealand GRAEME A. TAYLOR Research & Development Group, Department of Conservation, P.O. Box 10420, Wellington 6143, New Zealand Abstract An appraisal of the conservation status of the post-1800 New Zealand avifauna is presented. The list comprises 428 taxa in the following categories: ‘Extinct’ 20, ‘Threatened’ 77 (comprising 24 ‘Nationally Critical’, 15 ‘Nationally Endangered’, 38 ‘Nationally Vulnerable’), ‘At Risk’ 93 (comprising 18 ‘Declining’, 10 ‘Recovering’, 17 ‘Relict’, 48 ‘Naturally Uncommon’), ‘Not Threatened’ (native and resident) 36, ‘Coloniser’ 8, ‘Migrant’ 27, ‘Vagrant’ 130, and ‘Introduced and Naturalised’ 36. One species was assessed as ‘Data Deficient’. The list uses the New Zealand Threat Classification System, which provides greater resolution of naturally uncommon taxa typical of insular environments than the IUCN threat ranking system. New Zealand taxa are here ranked at subspecies level, and in some cases population level, when populations are judged to be potentially taxonomically distinct on the basis of genetic data or morphological observations. -

Ecuador Birding the Chocó-Andes Region: Western and Eastern Slopes of Ecuador November 25-December 4, 2018
MINDO CLOUD FOREST ECUADOR Birding the Chocó-Andes Region: Western and Eastern Slopes of Ecuador NOVEMBER 25-DECEMBER 4, 2018 Per square mile, Ecuador has the PROGRAM HIGHLIGHTS highest biodiversity in the world, including some 1,640 species of birds, • Explore diverse habitats at varying elevations at national and private reserves, including Alambi Cloud Forest Reserve, many of which are rare and endemic. the subtropical rainforest at Milpe Bird Sanctuary, páramo Discover the amazing contrasts of ecosystem at Antisana Ecological Reserve, and more. cloud forest, the Amazon, and Andean landscapes on this 10-day birding • Seek out some of Ecuador’s approximately 130 adventure. With assistance from an hummingbird species, including the Giant Hummingbird, Black-tailed Trainbearer, Sword-billed Hummingbird, expert guide, encounter a variety of Tourmaline Sunangel, and Glowing Puffleg. birds, plants, and other wildlife while traversing a selection of Ecuador’s • Identify other target species like the Andean Condor, 50 different ecosystems. The program Andean Cock-of-the-rock, Long-wattled Umbrellabird, Chocó Trogon, and dozens of tanagers. begins with a journey to Mindo Loma Private Reserve and its 17 acres of primary and secondary recovering cloud forest. Also experience the WHAt’s INCLUDED? Milpe Bird Sanctuary in the Chocó • Bilingual local guide Andean foothills, considered one of • Driver the finest sites in all of Ecuador. Along • Accommodations the way, meet a famous Ecuadorian • Activities ornithologist, view the snow-capped • Private transportation Antisana Volcano, and straddle the • Meals equator at the Middle of the World • Beverages with meals Monument. • Carbon offsetting BUFF-TAILED CORONET BY JULIAN LONDONO holbrooktravel.com | 800-451-7111 ANTISANA VOLCANO BY MARCIO RAMALHO ITINERARY areas of original cloud forest habitat and wildlife, where you will be able to experience the natural beauty and BLD = BREAKFAST, LUNCH, DINNER wonder of this very unique and biologically diverse environment.