Iverson, J.B. 1998. Molecules, Morphology, and Mud Turtle
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Ecology and Evolutionary History of Two Musk Turtles in the Southeastern United States
The University of Southern Mississippi The Aquila Digital Community Dissertations Spring 2020 The Ecology and Evolutionary History of Two Musk Turtles in the Southeastern United States Grover Brown Follow this and additional works at: https://aquila.usm.edu/dissertations Part of the Genetics Commons Recommended Citation Brown, Grover, "The Ecology and Evolutionary History of Two Musk Turtles in the Southeastern United States" (2020). Dissertations. 1762. https://aquila.usm.edu/dissertations/1762 This Dissertation is brought to you for free and open access by The Aquila Digital Community. It has been accepted for inclusion in Dissertations by an authorized administrator of The Aquila Digital Community. For more information, please contact [email protected]. THE ECOLOGY AND EVOLUTIONARY HISTORY OF TWO MUSK TURTLES IN THE SOUTHEASTERN UNITED STATES by Grover James Brown III A Dissertation Submitted to the Graduate School, the College of Arts and Sciences and the School of Biological, Environmental, and Earth Sciences at The University of Southern Mississippi in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy Approved by: Brian R. Kreiser, Committee Co-Chair Carl P. Qualls, Committee Co-Chair Jacob F. Schaefer Micheal A. Davis Willian W. Selman II ____________________ ____________________ ____________________ Dr. Brian R. Kreiser Dr. Jacob Schaefer Dr. Karen S. Coats Committee Chair Director of School Dean of the Graduate School May 2020 COPYRIGHT BY Grover James Brown III 2020 Published by the Graduate School ABSTRACT Turtles are among one of the most imperiled vertebrate groups on the planet with more than half of all species worldwide listed as threatened, endangered or extinct by the International Union of the Conservation of Nature. -
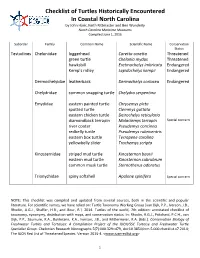
N.C. Turtles Checklist
Checklist of Turtles Historically Encountered In Coastal North Carolina by John Hairr, Keith Rittmaster and Ben Wunderly North Carolina Maritime Museums Compiled June 1, 2016 Suborder Family Common Name Scientific Name Conservation Status Testudines Cheloniidae loggerhead Caretta caretta Threatened green turtle Chelonia mydas Threatened hawksbill Eretmochelys imbricata Endangered Kemp’s ridley Lepidochelys kempii Endangered Dermochelyidae leatherback Dermochelys coriacea Endangered Chelydridae common snapping turtle Chelydra serpentina Emydidae eastern painted turtle Chrysemys picta spotted turtle Clemmys guttata eastern chicken turtle Deirochelys reticularia diamondback terrapin Malaclemys terrapin Special concern river cooter Pseudemys concinna redbelly turtle Pseudemys rubriventris eastern box turtle Terrapene carolina yellowbelly slider Trachemys scripta Kinosternidae striped mud turtle Kinosternon baurii eastern mud turtle Kinosternon subrubrum common musk turtle Sternotherus odoratus Trionychidae spiny softshell Apalone spinifera Special concern NOTE: This checklist was compiled and updated from several sources, both in the scientific and popular literature. For scientific names, we have relied on: Turtle Taxonomy Working Group [van Dijk, P.P., Iverson, J.B., Rhodin, A.G.J., Shaffer, H.B., and Bour, R.]. 2014. Turtles of the world, 7th edition: annotated checklist of taxonomy, synonymy, distribution with maps, and conservation status. In: Rhodin, A.G.J., Pritchard, P.C.H., van Dijk, P.P., Saumure, R.A., Buhlmann, K.A., Iverson, J.B., and Mittermeier, R.A. (Eds.). Conservation Biology of Freshwater Turtles and Tortoises: A Compilation Project of the IUCN/SSC Tortoise and Freshwater Turtle Specialist Group. Chelonian Research Monographs 5(7):000.329–479, doi:10.3854/crm.5.000.checklist.v7.2014; The IUCN Red List of Threatened Species. -

In AR, FL, GA, IA, KY, LA, MO, OH, OK, SC, TN, and TX): Species in Red = Depleted to the Point They May Warrant Federal Endangered Species Act Listing
Southern and Midwestern Turtle Species Affected by Commercial Harvest (in AR, FL, GA, IA, KY, LA, MO, OH, OK, SC, TN, and TX): species in red = depleted to the point they may warrant federal Endangered Species Act listing Common snapping turtle (Chelydra serpentina) – AR, GA, IA, KY, MO, OH, OK, SC, TX Florida common snapping turtle (Chelydra serpentina osceola) - FL Southern painted turtle (Chrysemys dorsalis) – AR Western painted turtle (Chrysemys picta) – IA, MO, OH, OK Spotted turtle (Clemmys gutatta) - FL, GA, OH Florida chicken turtle (Deirochelys reticularia chrysea) – FL Western chicken turtle (Deirochelys reticularia miaria) – AR, FL, GA, KY, MO, OK, TN, TX Barbour’s map turtle (Graptemys barbouri) - FL, GA Cagle’s map turtle (Graptemys caglei) - TX Escambia map turtle (Graptemys ernsti) – FL Common map turtle (Graptemys geographica) – AR, GA, OH, OK Ouachita map turtle (Graptemys ouachitensis) – AR, GA, OH, OK, TX Sabine map turtle (Graptemys ouachitensis sabinensis) – TX False map turtle (Graptemys pseudogeographica) – MO, OK, TX Mississippi map turtle (Graptemys pseuogeographica kohnii) – AR, TX Alabama map turtle (Graptemys pulchra) – GA Texas map turtle (Graptemys versa) - TX Striped mud turtle (Kinosternon baurii) – FL, GA, SC Yellow mud turtle (Kinosternon flavescens) – OK, TX Common mud turtle (Kinosternon subrubrum) – AR, FL, GA, OK, TX Alligator snapping turtle (Macrochelys temminckii) – AR, FL, GA, LA, MO, TX Diamond-back terrapin (Malaclemys terrapin) – FL, GA, LA, SC, TX River cooter (Pseudemys concinna) – AR, FL, -

The Staurotypus Turtles and Aves Share the Same Origin of Sex Chromosomes but Evolved Different Types of Heterogametic Sex Determination
The Staurotypus Turtles and Aves Share the Same Origin of Sex Chromosomes but Evolved Different Types of Heterogametic Sex Determination Taiki Kawagoshi1, Yoshinobu Uno1, Chizuko Nishida2, Yoichi Matsuda1,3* 1 Laboratory of Animal Genetics, Department of Applied Molecular Biosciences, Graduate School of Bioagricultural Sciences, Nagoya University, Nagoya, Japan, 2 Department of Natural History Sciences, Faculty of Science, Hokkaido University, Sapporo, Japan, 3 Avian Bioscience Research Center, Graduate School of Bioagricultural Sciences, Nagoya University, Nagoya, Japan Abstract Reptiles have a wide diversity of sex-determining mechanisms and types of sex chromosomes. Turtles exhibit temperature- dependent sex determination and genotypic sex determination, with male heterogametic (XX/XY) and female heterogametic (ZZ/ZW) sex chromosomes. Identification of sex chromosomes in many turtle species and their comparative genomic analysis are of great significance to understand the evolutionary processes of sex determination and sex chromosome differentiation in Testudines. The Mexican giant musk turtle (Staurotypus triporcatus, Kinosternidae, Testudines) and the giant musk turtle (Staurotypus salvinii) have heteromorphic XY sex chromosomes with a low degree of morphological differentiation; however, their origin and linkage group are still unknown. Cross-species chromosome painting with chromosome-specific DNA from Chinese soft-shelled turtle (Pelodiscus sinensis) revealed that the X and Y chromosomes of S. triporcatus have homology with P. sinensis chromosome 6, which corresponds to the chicken Z chromosome. We cloned cDNA fragments of S. triporcatus homologs of 16 chicken Z-linked genes and mapped them to S. triporcatus and S. salvinii chromosomes using fluorescence in situ hybridization. Sixteen genes were localized to the X and Y long arms in the same order in both species. -

Kinosternon Subrubrum (Bonnaterre 1789) – Eastern Mud Turtle
Conservation Biology of Freshwater Turtles and Tortoises: A Compilation ProjectKinosternidae of the IUCN SSC — KinosternonTortoise and Freshwater subrubrum Turtle Specialist Group 101.1 A.G.J. Rhodin, J.B. Iverson, P.P. van Dijk, K.A. Buhlmann, P.C.H. Pritchard, and R.A. Mittermeier, Eds. Chelonian Research Monographs (ISSN 1088-7105) No. 5, doi:10.3854/crm.5.101.subrubrum.v1.2017 © 2017 by Chelonian Research Foundation and Turtle Conservancy • Published 17 September 2017 Kinosternon subrubrum (Bonnaterre 1789) – Eastern Mud Turtle WALTER E. MESHAKA, JR.1, J. WHITFIELD GIBBONS2, DANIEL F. HUGHES3, MICHAEL W. KLEMENS4, AND JOHN B. IVERSON5 1State Museum of Pennsylvania, 300 North Street, Harrisburg, Pennsylvania 17120 USA [[email protected]]; 2Savannah River Ecology Lab, Drawer E, Aiken, South Carolina 29802 USA [[email protected]]; 3University of Texas at El Paso, El Paso, Texas 79968 USA [[email protected]]; 4Department of Herpetology, American Museum of Natural History, Central Park West at 79th Street, New York, New York 10024 USA [[email protected]]; 5Department of Biology, Earlham College, Richmond, Indiana 47374 USA [[email protected]] SUMMARY. — The Eastern Mud Turtle, Kinosternon subrubrum (Family Kinosternidae), is a small (carapace length 85 to 120 mm) polytypic species of the eastern and central United States. All three historically recognized subspecies (K. s. subrubrum, K. s. steindachneri, and K. s. hippocrepis) are semi-aquatic turtles that inhabit much of the U.S. Atlantic and Gulf Coastal Plains. The Florida taxon (K. s. steindachneri) appears to represent a distinct species, but we continue to treat it as a subspecies for the purposes of this account. -

Taxonomy and Phylogeny of the Higher Categories of Cryptodiran Turtles Based on a Cladistic Analysis of Chromosomal Data
Copein, 1983(4), pp. 918-932 Taxonomy and Phylogeny of the Higher Categories of Cryptodiran Turtles Based on a Cladistic Analysis of Chromosomal Data John W. Bickham and John L. Carr Karyological data are available for 55% of all cryptodiran turtle species including members of all but one family. Cladistic analysis of these data, as well as con sideration of other taxonomic studies, lead us to propose a formal classification and phylogeny not greatly different from that suggested by other workers. We recognize 11 families and three superfamilies. The platysternid and staurotypid turtles are recognized at the familial level. Patterns and models of karyotypic evolution in turtles are reviewed and discussed. OVER the past 10 years knowledge of turtle and the relationship between the shell and pel karyology has grown to such an extent vic girdle. In the cryptodires ("hidden-necked" that the order Testudines is one of the better turtles), the neck is withdrawn into the body in known groups of lower vertebrates (Bickham, a vertical plane and the pelvis is not fused to 1983). Nondifferentially stained karyotypes are either the plastron or carapace, whereas in the known for 55% of cryptodiran turtle species pleurodires ("side-necked" turtles) the pelvic and banded karyotypes for approximately 25% girdle is fused to both the plastron and carapace (Bickham, 1981). From this body of knowledge, and the neck is folded back against the body in as well as a consideration of the morphological a horizontal plane. Cope's suborder Athecae variation in the order, we herein present a gen includes only the Dermochelyidae and is no eral review of the cryptodiran karyological lit longer recognized. -

Proposed Amendment to 21CFR124021
Richard Fife 8195 S. Valley Vista Drive Hereford, AZ 85615 December 07, 2015 Division of Dockets Management Food and Drug Administration 5630 Fishers Lane, rm. 1061 Rockville, MD 20852 Reference: Docket Number FDA-2013-S-0610 Proposed Amendment to Code of Federal Regulations Title 21, Volume 8 Revised as of April 1, 2015 21CFR Sec.1240.62 Dear Dr. Stephen Ostroff, M.D., Acting Commissioner: Per discussion with the Division of Dockets Management staff on November 10, 2015 Environmental and Economic impact statements are not required for petitions submitted under 21CFR Sec.1240.62 CITIZEN PETITION December 07, 2015 ACTION REQUESTED: I propose an amendment to 21CFR Sec.1240.62 (see exhibit 1) as allowed by Section (d) Petitions as follows: Amend section (c) Exceptions. The provisions of this section are not applicable to: By adding the following two (2) exceptions: (5) The sale, holding for sale, and distribution of live turtles and viable turtle eggs, which are sold for a retail value of $75 or more (not to include any additional turtle related apparatuses, supplies, cages, food, or other turtle related paraphernalia). This dollar amount should be reviewed every 5 years or more often, as deemed necessary by the department in order to make adjustments for inflation using the US Department of Labor, Bureau of labor Statistics, Consumer Price Index. (6) The sale, holding for sale, and distribution of live turtles and viable turtle eggs, which are listed by the International Union for Conservation of Nature and Natural Resources (IUCN) Red List as Extinct In Wild, Critically Endangered, Endangered, or Vulnerable (IUCN threatened categorizes). -

Heinrich and Walsh (2019)
THE BIG TURTLE YEAR LOOKING FOR WILD TURTLES IN WILD PLACES By George L. Heinrich and Timothy J. Walsh e like looking for wild turtles in wild We both liked turtles as kids, but now, many places. From the time George was in years later, we understand the important ecological Welementary school catching Wood roles they play. Some turtle species serve as indica- Turtles in southwestern Connecticut and Tim tors of environmental health, while others are clas- found his first pebble-sized Striped Mud Turtle sified as keystone species (those that play a vital at age 10 in a south Florida stream, we have both ecological role in a given habitat), umbrella spe- marveled at being in nature and searching for these cies (those whose conservation benefits the larger fascinating reptiles. As children, little did we know ecological community), or flagship species (iconic that our time spent exploring our neighborhood symbols of habitat conservation efforts). Perhaps woods would lead to rewarding careers in wildlife one of the best examples of the ecological signifi- conservation. Now we are both officers with the cance of turtles is the Gopher Tortoise, an imper- Florida Turtle Conservation Trust, an organization iled species that occurs in a six-state range within working to conserve Florida’s rich turtle diversity. the Southeastern Coastal Plain of the United States. In 2017 we created the opportunity of a Appropriately, the Gopher Tortoise was the first lifetime with the Trust’s conservation education species we found on day one of The Big Turtle Year. initiative: The Big Turtle Year, an ambitious plan to travel across the United States and back again SoutheaSt Regional highlightS trying to find as many species as we could. -

Water Turtles
WATER TURTLES Range Bog Turtle Bog Turtle • North America’s smallest turtle (3” to 4.5”); NC’s rarest • Have noticeable bright orange, yellow or red blotch on each side of face • Live in isolated spring-fed fen, sphagnum bogs, marshy meadows and wet pastures • Eat beetles, insect larvae, snails, seeds and millipedes • Females mature from 5-8 years and mate from May to Yellow-bellied Slider June Range • Deposit 2-6 eggs from June to July which hatch after 42 -56 days of incubation • Placed on NC’s threatened list in 1989 ENDANGERED 1 Yellow-bellied Slider • Yellow patch on side of head – on females and juveniles • 5 to 8 inches • Underside of shell is yellow • Males smaller than females with longer, thicker tail and long fingernails • Juveniles eat more carnivorous diet Redbelly Turtle • Adults are omnivores – feeding = underwater • Males mature between 3-5 yrs • 5-7 year old females lay 4 to 23 eggs from May to July • Eggs incubate 2-2.5 months and hatch between July- September • Help control invertebrate and vegetation populations • Live in fresh water Range Red Belly Turtle • 10-15” in length • Like deep water • Eat both plants and animals-insect larvae, crayfish, worms and tadpoles • Active from May thru October Snapping Turtle • Females lay 10-12 eggs in June-July which hatch Range sometime in late summer. • Young sometimes winter over in the nest until spring • Like to bask in the sun 2 Snapping Turtle • 8-14 inches weighing from 10 to 50 lbs • Large head, small plastron, long tail & strong limbs • Males larger than females Range -

Chelonian Advisory Group Regional Collection Plan 4Th Edition December 2015
Association of Zoos and Aquariums (AZA) Chelonian Advisory Group Regional Collection Plan 4th Edition December 2015 Editor Chelonian TAG Steering Committee 1 TABLE OF CONTENTS Introduction Mission ...................................................................................................................................... 3 Steering Committee Structure ........................................................................................................... 3 Officers, Steering Committee Members, and Advisors ..................................................................... 4 Taxonomic Scope ............................................................................................................................. 6 Space Analysis Space .......................................................................................................................................... 6 Survey ........................................................................................................................................ 6 Current and Potential Holding Table Results ............................................................................. 8 Species Selection Process Process ..................................................................................................................................... 11 Decision Tree ........................................................................................................................... 13 Decision Tree Results ............................................................................................................. -

The Karyotype and Chromosomal Banding Patterns of the Central American River Turtle Dermatemys Mawii
Herpetologica, 37(2), 1981, 92-95 © 1981 by The Herpetologists' League, Inc. THE KARYOTYPE AND CHROMOSOMAL BANDING PATTERNS OF THE CENTRAL AMERICAN RIVER TURTLE DERMATEMYS MAWII John L. Carr, John W. Bickham, and Robert H. Dean Abstract: Dermatemys mawii has a diploid number of 56 and a karyotype identical to that of a sea turtle (Chelonia mydas). We hypothesize that D. mawii and C. mydas share the primitive karyotype for all non-trionychoid cryptodiran turtles. Key words: Cryptodira; Dermatemydidae; Dermatemys; Karyology; Reptilia; Testudines The family Dermatemydidae is one of The only living dermatemydid is Der- the oldest extant families of the suborder matemys mawii, a large river turtle from Cryptodira, with a fossil record extending southern Mexico and adjacent Central back into the Cretaceous (Gaffney, 1975). America (Iverson and Mittermeier, 1980). June 1981] HERPETOLOGICA 93 |l II 19 ti it mm B II II II «• mm* •■ mm FlG. 1.—Standard karyotype of Dermatemys mawii with chromosomes arranged into groups: (A) meta- centric to submetacentric macrochromosomes, (B) telocentric to subtelocentric macrochromosomes, and (C) microchromosomes. Dermatemys has been closely allied to in Tuxtla Gutierrez, Chiapas, Mexico. the mud and musk turtles of the family Exact locality data were unavailable, but Kinosternidae on the basis of serum pro the specimen was presumably from the tein electrophoresis (Frair, 1964, 1972), state of Chiapas. cranial morphology (Gaffhey, 1975), and penial morphology (Zug, 1966). Based Results upon pelvic girdle and hindlimb mor phology, Zug (1971) further proposed The standard karyotype of D. mawii that Dermatemys represents the basal (2n = 56) is presented in Figure 1. Chro stock of the kinosternids. -

Turtle Taxonomy Working Group 2007B
TURTLE TAXONOMY WORKING GROUP – Annotated List of Turtle Taxa 173 Defining Turtle Diversity: Proceedings of a Workshop on Genetics, Ethics, and Taxonomy of Freshwater Turtles and Tortoises H. Bradley Shaffer, Nancy N. FitzSimmons, Arthur Georges, and Anders G.J. Rhodin, Eds. Chelonian Research Monographs 4:173–199 • © 2007 by Chelonian Research Foundation An Annotated List of Modern Turtle Terminal Taxa with Comments on Areas of Taxonomic Instability and Recent Change TURTLE TAXONOMY WORKING GROUP* *Authorship of this article is by this group, which for the purposes of this document consisted of the following contributors listed alphabetically: JOHN W. BICKHAM1, JOHN B. IVERSON2, JAMES F. PARHAM3*, HANS-DIETER PHILIPPEN4, ANDERS G.J. RHODIN5*, H. BRADLEY SHAFFER6, PHILLIP Q. SPINKS6, AND PETER PAUL VAN DIJK7 1Center for the Environment, Purdue University, 503 Northwestern Avenue, West Lafayette, IN 47907 USA [[email protected]]; 2Department of Biology, Earlham college, Richmond, IN 47374 USA [[email protected]]; 3Department of Herpetology, California Academy of Sciences, 875 Howard Street, San Francisco, CA 94103 USA, and Museum of Paleontology, 1101 Valley Life Sciences Building, University of California, Berkeley, CA 94720 USA [[email protected]]; 4Hans-Dieter Philippen, Kuhlertstrasse 154, D-52525 Heinsberg, Germany [[email protected]]; 5Chelonian Research Foundation, 168 Goodrich Street, Lunenburg, MA 01462 USA [[email protected]]; 6Section of Evolution and Ecology, and Center for Population Biology, University of California, Davis, CA 95616 USA [[email protected], [email protected]]; 7CI/CABS Tortoise and Freshwater Turtle Conservation Program, Center for Applied Biodiversity Science, Conservation International, 2011 Crystal Drive, Suite 500, Arlington, VA 22202 USA [[email protected]]; *Corresponding authors and primary collators ABSTRACT.