Taxonomy and Phylogeny of the Higher Categories of Cryptodiran Turtles Based on a Cladistic Analysis of Chromosomal Data
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Ecology and Evolutionary History of Two Musk Turtles in the Southeastern United States
The University of Southern Mississippi The Aquila Digital Community Dissertations Spring 2020 The Ecology and Evolutionary History of Two Musk Turtles in the Southeastern United States Grover Brown Follow this and additional works at: https://aquila.usm.edu/dissertations Part of the Genetics Commons Recommended Citation Brown, Grover, "The Ecology and Evolutionary History of Two Musk Turtles in the Southeastern United States" (2020). Dissertations. 1762. https://aquila.usm.edu/dissertations/1762 This Dissertation is brought to you for free and open access by The Aquila Digital Community. It has been accepted for inclusion in Dissertations by an authorized administrator of The Aquila Digital Community. For more information, please contact [email protected]. THE ECOLOGY AND EVOLUTIONARY HISTORY OF TWO MUSK TURTLES IN THE SOUTHEASTERN UNITED STATES by Grover James Brown III A Dissertation Submitted to the Graduate School, the College of Arts and Sciences and the School of Biological, Environmental, and Earth Sciences at The University of Southern Mississippi in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy Approved by: Brian R. Kreiser, Committee Co-Chair Carl P. Qualls, Committee Co-Chair Jacob F. Schaefer Micheal A. Davis Willian W. Selman II ____________________ ____________________ ____________________ Dr. Brian R. Kreiser Dr. Jacob Schaefer Dr. Karen S. Coats Committee Chair Director of School Dean of the Graduate School May 2020 COPYRIGHT BY Grover James Brown III 2020 Published by the Graduate School ABSTRACT Turtles are among one of the most imperiled vertebrate groups on the planet with more than half of all species worldwide listed as threatened, endangered or extinct by the International Union of the Conservation of Nature. -

Egyptian Tortoise (Testudo Kleinmanni)
EAZA Reptile Taxon Advisory Group Best Practice Guidelines for the Egyptian tortoise (Testudo kleinmanni) First edition, May 2019 Editors: Mark de Boer, Lotte Jansen & Job Stumpel EAZA Reptile TAG chair: Ivan Rehak, Prague Zoo. EAZA Best Practice Guidelines Egyptian tortoise (Testudo kleinmanni) EAZA Best Practice Guidelines disclaimer Copyright (May 2019) by EAZA Executive Office, Amsterdam. All rights reserved. No part of this publication may be reproduced in hard copy, machine-readable or other forms without advance written permission from the European Association of Zoos and Aquaria (EAZA). Members of the European Association of Zoos and Aquaria (EAZA) may copy this information for their own use as needed. The information contained in these EAZA Best Practice Guidelines has been obtained from numerous sources believed to be reliable. EAZA and the EAZA Reptile TAG make a diligent effort to provide a complete and accurate representation of the data in its reports, publications, and services. However, EAZA does not guarantee the accuracy, adequacy, or completeness of any information. EAZA disclaims all liability for errors or omissions that may exist and shall not be liable for any incidental, consequential, or other damages (whether resulting from negligence or otherwise) including, without limitation, exemplary damages or lost profits arising out of or in connection with the use of this publication. Because the technical information provided in the EAZA Best Practice Guidelines can easily be misread or misinterpreted unless properly analysed, EAZA strongly recommends that users of this information consult with the editors in all matters related to data analysis and interpretation. EAZA Preamble Right from the very beginning it has been the concern of EAZA and the EEPs to encourage and promote the highest possible standards for husbandry of zoo and aquarium animals. -
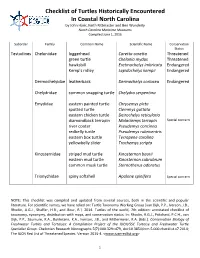
N.C. Turtles Checklist
Checklist of Turtles Historically Encountered In Coastal North Carolina by John Hairr, Keith Rittmaster and Ben Wunderly North Carolina Maritime Museums Compiled June 1, 2016 Suborder Family Common Name Scientific Name Conservation Status Testudines Cheloniidae loggerhead Caretta caretta Threatened green turtle Chelonia mydas Threatened hawksbill Eretmochelys imbricata Endangered Kemp’s ridley Lepidochelys kempii Endangered Dermochelyidae leatherback Dermochelys coriacea Endangered Chelydridae common snapping turtle Chelydra serpentina Emydidae eastern painted turtle Chrysemys picta spotted turtle Clemmys guttata eastern chicken turtle Deirochelys reticularia diamondback terrapin Malaclemys terrapin Special concern river cooter Pseudemys concinna redbelly turtle Pseudemys rubriventris eastern box turtle Terrapene carolina yellowbelly slider Trachemys scripta Kinosternidae striped mud turtle Kinosternon baurii eastern mud turtle Kinosternon subrubrum common musk turtle Sternotherus odoratus Trionychidae spiny softshell Apalone spinifera Special concern NOTE: This checklist was compiled and updated from several sources, both in the scientific and popular literature. For scientific names, we have relied on: Turtle Taxonomy Working Group [van Dijk, P.P., Iverson, J.B., Rhodin, A.G.J., Shaffer, H.B., and Bour, R.]. 2014. Turtles of the world, 7th edition: annotated checklist of taxonomy, synonymy, distribution with maps, and conservation status. In: Rhodin, A.G.J., Pritchard, P.C.H., van Dijk, P.P., Saumure, R.A., Buhlmann, K.A., Iverson, J.B., and Mittermeier, R.A. (Eds.). Conservation Biology of Freshwater Turtles and Tortoises: A Compilation Project of the IUCN/SSC Tortoise and Freshwater Turtle Specialist Group. Chelonian Research Monographs 5(7):000.329–479, doi:10.3854/crm.5.000.checklist.v7.2014; The IUCN Red List of Threatened Species. -

A Phylogenomic Analysis of Turtles ⇑ Nicholas G
Molecular Phylogenetics and Evolution 83 (2015) 250–257 Contents lists available at ScienceDirect Molecular Phylogenetics and Evolution journal homepage: www.elsevier.com/locate/ympev A phylogenomic analysis of turtles ⇑ Nicholas G. Crawford a,b,1, James F. Parham c, ,1, Anna B. Sellas a, Brant C. Faircloth d, Travis C. Glenn e, Theodore J. Papenfuss f, James B. Henderson a, Madison H. Hansen a,g, W. Brian Simison a a Center for Comparative Genomics, California Academy of Sciences, 55 Music Concourse Drive, San Francisco, CA 94118, USA b Department of Genetics, University of Pennsylvania, Philadelphia, PA 19104, USA c John D. Cooper Archaeological and Paleontological Center, Department of Geological Sciences, California State University, Fullerton, CA 92834, USA d Department of Biological Sciences, Louisiana State University, Baton Rouge, LA 70803, USA e Department of Environmental Health Science, University of Georgia, Athens, GA 30602, USA f Museum of Vertebrate Zoology, University of California, Berkeley, CA 94720, USA g Mathematical and Computational Biology Department, Harvey Mudd College, 301 Platt Boulevard, Claremont, CA 9171, USA article info abstract Article history: Molecular analyses of turtle relationships have overturned prevailing morphological hypotheses and Received 11 July 2014 prompted the development of a new taxonomy. Here we provide the first genome-scale analysis of turtle Revised 16 October 2014 phylogeny. We sequenced 2381 ultraconserved element (UCE) loci representing a total of 1,718,154 bp of Accepted 28 October 2014 aligned sequence. Our sampling includes 32 turtle taxa representing all 14 recognized turtle families and Available online 4 November 2014 an additional six outgroups. Maximum likelihood, Bayesian, and species tree methods produce a single resolved phylogeny. -

The Conservation Biology of Tortoises
The Conservation Biology of Tortoises Edited by Ian R. Swingland and Michael W. Klemens IUCN/SSC Tortoise and Freshwater Turtle Specialist Group and The Durrell Institute of Conservation and Ecology Occasional Papers of the IUCN Species Survival Commission (SSC) No. 5 IUCN—The World Conservation Union IUCN Species Survival Commission Role of the SSC 3. To cooperate with the World Conservation Monitoring Centre (WCMC) The Species Survival Commission (SSC) is IUCN's primary source of the in developing and evaluating a data base on the status of and trade in wild scientific and technical information required for the maintenance of biological flora and fauna, and to provide policy guidance to WCMC. diversity through the conservation of endangered and vulnerable species of 4. To provide advice, information, and expertise to the Secretariat of the fauna and flora, whilst recommending and promoting measures for their con- Convention on International Trade in Endangered Species of Wild Fauna servation, and for the management of other species of conservation concern. and Flora (CITES) and other international agreements affecting conser- Its objective is to mobilize action to prevent the extinction of species, sub- vation of species or biological diversity. species, and discrete populations of fauna and flora, thereby not only maintain- 5. To carry out specific tasks on behalf of the Union, including: ing biological diversity but improving the status of endangered and vulnerable species. • coordination of a programme of activities for the conservation of biological diversity within the framework of the IUCN Conserva- tion Programme. Objectives of the SSC • promotion of the maintenance of biological diversity by monitor- 1. -

A Case of Inversion and Duplication Involving Constitutive Heterochromatin
Genetics and Molecular Biology, 36, 3, 353-356 (2013) Copyright © 2013, Sociedade Brasileira de Genética. Printed in Brazil www.sbg.org.br Short Communication Cytogenetic comparison of Podocnemis expansa and Podocnemis unifilis: A case of inversion and duplication involving constitutive heterochromatin Ricardo José Gunski1, Isabel Souza Cunha2, Tiago Marafiga Degrandi1, Mario Ledesma3 and Analía Del Valle Garnero1 1Universidade Federal do Pampa, Campus São Gabriel, São Gabriel, RS, Brazil. 2Secretaria Municipal de Meio Ambiente e Turismo, São Desidério, BA, Brazil. 3Parque Ecológico “El Puma”, Candelaria, Argentina. Abstract Podocnemis expansa and P. unifilis present 2n = 28 chromosomes, a diploid number similar to those observed in other species of the genus. The aim of this study was to characterize these two species using conventional staining and differential CBG-, GTG and Ag-NOR banding. We analyzed specimens of P. expansa and P. unifilis from the state of Tocantins (Brazil), in which we found a 2n = 28 and karyotypes differing in the morphology of the 13th pair, which was submetacentric in P. expansa and telocentric in P. unifilis. The CBG-banding patterns revealed a heterochromatic block in the short arm of pair 13 of P. expansa and an interstitial one in pair 13 of P. unifilis, suggest- ing a pericentric inversion. Pair 14 of P. unifilis showed an insterstitial band in the long arm that was absent in P. expansa, suggesting a duplication in this region. Ag-NORs were observed in the first chromosome pair of both spe- cies and was associated to a secondary constriction and heterochromatic blocks. Keywords: chromosome banding, Ag-NORs, karyotype, chelonids. -

Dermatemys Mawii (The Hicatee, Tortuga Blanca, Or Central American River Turtle): a Working Bibliography
See discussions, stats, and author profiles for this publication at: https://www.researchgate.net/publication/325206006 Dermatemys mawii (The Hicatee, Tortuga Blanca, or Central American River Turtle): A Working Bibliography Article · May 2018 CITATIONS READS 0 521 6 authors, including: Venetia Briggs Sergio C. Gonzalez University of Florida University of Florida 16 PUBLICATIONS 133 CITATIONS 9 PUBLICATIONS 39 CITATIONS SEE PROFILE SEE PROFILE Thomas Rainwater Clemson University 143 PUBLICATIONS 1,827 CITATIONS SEE PROFILE Some of the authors of this publication are also working on these related projects: Crocodiles in the Everglades View project Dissertation: The role of habitat expansion on amphibian community structure View project All content following this page was uploaded by Sergio C. Gonzalez on 17 May 2018. The user has requested enhancement of the downloaded file. 2018 Endangered and ThreatenedCaribbean Species Naturalist of the Caribbean Region Special Issue No. 2 V. Briggs-Gonzalez, S.C. Gonzalez, D. Smith, K. Allen, T.R. Rainwater, and F.J. Mazzotti 2018 CARIBBEAN NATURALIST Special Issue No. 2:1–22 Dermatemys mawii (The Hicatee, Tortuga Blanca, or Central American River Turtle): A Working Bibliography Venetia Briggs-Gonzalez1,*, Sergio C. Gonzalez1, Dustin Smith2, Kyle Allen1, Thomas R. Rainwater3, and Frank J. Mazzotti1 Abstract - Dermatemys mawii (Central American River Turtle), locally known in Belize as the “Hicatee” and in Guatemala and Mexico as Tortuga Blanca, is a large, highly aquatic freshwater turtle that has been extirpated from much of its historical range of southern Mex- ico, northern Guatemala, and lowland Belize. Throughout its restricted range, Dermatemys has been intensely harvested for its meat and eggs and sold in local markets. -

Diets of Freshwater Turtles Often Reflect the Availability of Food Resources in the Environment
Herpetological Conservation and Biology 8(3):561−570. HerpetologicalSubmitted: 26 March Conservation 2013; Accepted: and Biology 21 October 2013; Published: 31 December 2013. RazoR-Backed Musk TuRTle (SternotheruS carinatuS) dieT acRoss a GRadienT of invasion carla l. atkinSon1,2, 3 1Oklahoma Biological Survey, 111 E. Chesapeake St., Norman, OK 73019 2Department of Biology and Ecology and Evolutionary Biology Graduate Program, University of Oklahoma, Norman, Ok 73019 3Present Address: Dept. of Ecology and Evolutionary Biology, Corson Hall, Cornell University, Ithaca, NY 14853 e-mail: [email protected] abstract.—diets of freshwater turtles often reflect the availability of food resources in the environment. accordingly, bottom- feeding turtles’ diets are typically composed of benthic macroinvertebrate fauna (e.g., insects and mollusks). However, the composition of benthic systems has changed because many freshwater ecosystems have been invaded by non-native species, including bivalve species such as the asian clam, corbicula fluminea. i studied the diet of Sternotherus carinatus, the Razor- backed Musk Turtle, in southeastern oklahoma across three zones of corbicula abundances: no corbicula, moderate corbicula densities, and high corbicula densities. i hypothesized that the composition of corbicula in the diet would increase with increased abundance of corbicula in the riverine environment. Turtles were caught by snorkel surveys in the little and Mountain fork rivers and kept overnight for the collection of fecal samples. The diet was similar to that found in previous studies on S. carinatus except that corbicula is a new component of the diet and composed the majority of the diet in high-density corbicula areas. an index of Relative importance (iRi) showed that corbicula was the most important prey item in the areas with high corbicula density, was equally as important as gastropods in the areas with moderate corbicula density, and was absent from the diet in areas without corbicula. -

Turtles, All Marine Turtles, Have Been Documented Within the State’S Borders
Turtle Only four species of turtles, all marine turtles, have been documented within the state’s borders. Terrestrial and freshwater aquatic species of turtles do not occur in Alaska. Marine turtles are occasional visitors to Alaska’s Gulf Coast waters and are considered a natural part of the state’s marine ecosystem. Between 1960 and 2007 there were 19 reports of leatherback sea turtles (Dermochelys coriacea), the world’s largest turtle. There have been 15 reports of Green sea turtles (Chelonia mydas). The other two are extremely rare, there have been three reports of Olive ridley sea turtles (Lepidochelys olivacea) and two reports of loggerhead sea turtles (Caretta caretta). Currently, all four species are listed as threatened or endangered under the U.S. Endangered Species Act. Prior to 1993, Alaska marine turtle sightings were mostly of live leatherback sea turtles; since then most observations have been of green sea turtle carcasses. At present, it is not possible to determine if this change is related to changes in oceanographic conditions, perhaps as the result of global warming, or to changes in the overall population size and distribution of these species. General description: Marine turtles are large, tropical/subtropical, thoroughly aquatic reptiles whose forelimbs or flippers are specially modified for swimming and are considerably larger than their hind limbs. Movements on land are awkward. Except for occasional basking by both sexes and egg-laying by females, turtles rarely come ashore. Turtles are among the longest-lived vertebrates. Although their age is often exaggerated, they probably live 50 to 100 years. Of the five recognized species of marine turtles, four (including the green sea turtle) belong to the family Cheloniidae. -

Mitochondrial Genome Reveals Contrasting Pattern of Adaptive
bioRxiv preprint doi: https://doi.org/10.1101/2021.02.18.431795; this version posted February 18, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-ND 4.0 International license. MITOCHONDRIAL GENOME REVEALS CONTRASTING PATTERN OF ADAPTIVE SELECTION IN TURTLES AND TORTOISES Subhashree Sahoo1,2, Ajit Kumar1, Jagdish Rai2, Sandeep Kumar Gupta1 1. Department of Animal Ecology and Conservation Biology, Wildlife Institute of India, Dehradun 2. Institute of Forensic Science and Criminology, Panjab University, India *Author for correspondence Dr. S.K. Gupta Scientist-E Wildlife Institute of India Dehra Dun 248 001, India E-mail: [email protected] Telephone: +91-135-2646343 Fax No.: +91-135-2640117 1 bioRxiv preprint doi: https://doi.org/10.1101/2021.02.18.431795; this version posted February 18, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-ND 4.0 International license. Abstract Testudinoidea represents an evolutionarily unique taxon comprising both turtles and tortoises. The contrasting habitats that turtles and tortoises inhabit are associated with unique physio-ecological challenges hence enable distinct adaptive evolutionary strategies. To comparatively understand the pattern and strength of Darwinian selection and physicochemical evolution in turtle and tortoise mitogenomes, we employed adaptive divergence and selection analyses. We evaluated changes in structural and biochemical properties, and codon models on the mitochondrial protein-coding genes (PCGs) among three turtles and a tortoise lineage. -

In AR, FL, GA, IA, KY, LA, MO, OH, OK, SC, TN, and TX): Species in Red = Depleted to the Point They May Warrant Federal Endangered Species Act Listing
Southern and Midwestern Turtle Species Affected by Commercial Harvest (in AR, FL, GA, IA, KY, LA, MO, OH, OK, SC, TN, and TX): species in red = depleted to the point they may warrant federal Endangered Species Act listing Common snapping turtle (Chelydra serpentina) – AR, GA, IA, KY, MO, OH, OK, SC, TX Florida common snapping turtle (Chelydra serpentina osceola) - FL Southern painted turtle (Chrysemys dorsalis) – AR Western painted turtle (Chrysemys picta) – IA, MO, OH, OK Spotted turtle (Clemmys gutatta) - FL, GA, OH Florida chicken turtle (Deirochelys reticularia chrysea) – FL Western chicken turtle (Deirochelys reticularia miaria) – AR, FL, GA, KY, MO, OK, TN, TX Barbour’s map turtle (Graptemys barbouri) - FL, GA Cagle’s map turtle (Graptemys caglei) - TX Escambia map turtle (Graptemys ernsti) – FL Common map turtle (Graptemys geographica) – AR, GA, OH, OK Ouachita map turtle (Graptemys ouachitensis) – AR, GA, OH, OK, TX Sabine map turtle (Graptemys ouachitensis sabinensis) – TX False map turtle (Graptemys pseudogeographica) – MO, OK, TX Mississippi map turtle (Graptemys pseuogeographica kohnii) – AR, TX Alabama map turtle (Graptemys pulchra) – GA Texas map turtle (Graptemys versa) - TX Striped mud turtle (Kinosternon baurii) – FL, GA, SC Yellow mud turtle (Kinosternon flavescens) – OK, TX Common mud turtle (Kinosternon subrubrum) – AR, FL, GA, OK, TX Alligator snapping turtle (Macrochelys temminckii) – AR, FL, GA, LA, MO, TX Diamond-back terrapin (Malaclemys terrapin) – FL, GA, LA, SC, TX River cooter (Pseudemys concinna) – AR, FL, -

The Staurotypus Turtles and Aves Share the Same Origin of Sex Chromosomes but Evolved Different Types of Heterogametic Sex Determination
The Staurotypus Turtles and Aves Share the Same Origin of Sex Chromosomes but Evolved Different Types of Heterogametic Sex Determination Taiki Kawagoshi1, Yoshinobu Uno1, Chizuko Nishida2, Yoichi Matsuda1,3* 1 Laboratory of Animal Genetics, Department of Applied Molecular Biosciences, Graduate School of Bioagricultural Sciences, Nagoya University, Nagoya, Japan, 2 Department of Natural History Sciences, Faculty of Science, Hokkaido University, Sapporo, Japan, 3 Avian Bioscience Research Center, Graduate School of Bioagricultural Sciences, Nagoya University, Nagoya, Japan Abstract Reptiles have a wide diversity of sex-determining mechanisms and types of sex chromosomes. Turtles exhibit temperature- dependent sex determination and genotypic sex determination, with male heterogametic (XX/XY) and female heterogametic (ZZ/ZW) sex chromosomes. Identification of sex chromosomes in many turtle species and their comparative genomic analysis are of great significance to understand the evolutionary processes of sex determination and sex chromosome differentiation in Testudines. The Mexican giant musk turtle (Staurotypus triporcatus, Kinosternidae, Testudines) and the giant musk turtle (Staurotypus salvinii) have heteromorphic XY sex chromosomes with a low degree of morphological differentiation; however, their origin and linkage group are still unknown. Cross-species chromosome painting with chromosome-specific DNA from Chinese soft-shelled turtle (Pelodiscus sinensis) revealed that the X and Y chromosomes of S. triporcatus have homology with P. sinensis chromosome 6, which corresponds to the chicken Z chromosome. We cloned cDNA fragments of S. triporcatus homologs of 16 chicken Z-linked genes and mapped them to S. triporcatus and S. salvinii chromosomes using fluorescence in situ hybridization. Sixteen genes were localized to the X and Y long arms in the same order in both species.