Mcnab Genetic Study
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Beauceron
The Beauceron The Beauceron, also known as Berger de Beauce and Bas Rouge, is the largest of the French sheepdogs and was developed solely in France with no foreign crosses. The Beauceron is closely related to the longhaired Briard or Berger de Brie. The first mention of a dog which matches the Beauceron’s description is found in a manuscript dated 1587. In 1809 Abbé Rozier wrote an article on French herding dogs. It was he who first described the differences in type and used the terms Berger de la Brie for long coated dogs and Berger de la Beauce for short coated dogs. The name Beauceron was used for the first time by Pierre Megnin in his 1888 book on war dogs and the first Berger de Beauce was registered with the Societe Central Canine in September 1893. The French Club Les Amis du Beauceron (CAB), was founded in 1922 by Pierre Megnin and he together with Emmanuel Boulet developed the original breed standard for the Beauceron. The CAB has since guided the development of the breed in its native France, always keeping a watchful eye on the preservation of the breed’s herding and working ability. During the early part of the 19th century large flocks of sheep were common and the Beauceron was indispensable for the shepherds of France; two dogs were sufficient to tend to flocks of 200 to 300 head of sheep. Sheep production experienced a sharp decline during the later half of the 19th century and by the second half of the 20th century was only a phantom of its past. -

Frequently Asked Questions
APPLYING SHAMPOO & CONDITIONER For best results apply the shampoo & conditioner in the following order: 1. belly & Determine the appropriate shampoo & conditioner for your breed. chest 2. legs and feet 3. neck & face 4. back SHORT COATS need LEMON, MEDIUM COATS need BANANA and bottom. Let soak for 5 full minutes. These LONG COATS need GREEN APPLE. are the sebaceous areas where most dirt and toxins enter the pet’s system. SHAMPOO: Dilute 1 part shampoo to 3 parts water in a clean container. Wet the coat & apply shampoo. Soak for 5 full minutes. Rinse. CONDITIONER: Dilute 1 part conditioner to 3 parts water in a clean container. Apply conditioner to coat. Soak for 5 full minutes. Rinse. Don’t see your breed? Please visit Please visit our info site http://isbusa.com/ instructions/akc-breeds-by-coat-type/ Frequently Asked Questions: SHORT COATS MEDIUM COATS LONG COATS Why are there different products for different American Foxhound Bernese Mountain Dog Afghan Hound breeds? All pet coats need oils, minerals and Staffordshire Terrier Brussels Griffon Bearded Collie collagen to thrive. However short coats need Basenji Cairn Terrier Bedlington Terrier more oils, medium coats need more minerals Basset Hound Cavalier King Charles Bichon Frise and long coats need more collagen. These Beagle Chesapeake Bay Retriever Bolonese products are created to provide the right Beauceron Chow Chow Chinese Crested balance of each for all coat types. Bloodhound Cocker Spaniel Coton De Tulear Why do I have to bathe per your instructions? Boston Terrier German Shepherd Havanese Since pets absorb dirt & toxins from the ground Boxer Golden Retriever Japanese Chin up into their sebaceous areas (tummy, feet, Great Dane Irish Wolfhound Lhasa Apso legs and other areas with less hair) it is Bull Terrier Keeshond Longhaired Dachshund important that the products are applied there Chihuahua Labrador Retriever Maltese first & allowed to soak for 5 min. -

Dog Breeds of the World
Dog Breeds of the World Get your own copy of this book Visit: www.plexidors.com Call: 800-283-8045 Written by: Maria Sadowski PlexiDor Performance Pet Doors 4523 30th St West #E502 Bradenton, FL 34207 http://www.plexidors.com Dog Breeds of the World is written by Maria Sadowski Copyright @2015 by PlexiDor Performance Pet Doors Published in the United States of America August 2015 All rights reserved. No portion of this book may be reproduced or transmitted in any form or by any electronic or mechanical means, including photocopying, recording, or by any information retrieval and storage system without permission from PlexiDor Performance Pet Doors. Stock images from canstockphoto.com, istockphoto.com, and dreamstime.com Dog Breeds of the World It isn’t possible to put an exact number on the Does breed matter? dog breeds of the world, because many varieties can be recognized by one breed registration The breed matters to a certain extent. Many group but not by another. The World Canine people believe that dog breeds mostly have an Organization is the largest internationally impact on the outside of the dog, but through the accepted registry of dog breeds, and they have ages breeds have been created based on wanted more than 340 breeds. behaviors such as hunting and herding. Dog breeds aren’t scientifical classifications; they’re It is important to pick a dog that fits the family’s groupings based on similar characteristics of lifestyle. If you want a dog with a special look but appearance and behavior. Some breeds have the breed characterics seem difficult to handle you existed for thousands of years, and others are fairly might want to look for a mixed breed dog. -

Issue SC046 50Y056 March-April 2021
The Official Publicaon of The Sutherland Shire Dog Training Club Inc ABN: 85 904 409 654 Visit us at: www.ssdtc.com.au Issue SC046-50Y056 March-April 2021 Sutherland Shire Dog Training Club Inc . (Affiliated with the Dogs NSW) ABN :85 904 409 Postal Address: PO BOX 231 SUTHERLAND NSW 1499 (02) 9521 1633 (Clubhouse, during training hours) Web : www.ssdtc.com.au Email: [email protected] Your Committee 2020 - 2021 Other Positions President Pam Hazleton Assistant Treasurer Steve Burness Vice President Ron Brouwer Grounds Secretary Wendy Bourke Treasurer Elwyn Gaywood Grounds Secretary Andrea Munn Secretary Rob Collaro The Wag Editor Shanelle Corben Andrea Munn Instructors’ Facilitator Ros Klumpes Peter Stone Vetting Officer Rob Collaro Steve Burness Facility Officer Brian Jarrett Trial Secretary Margaret Taylor Agility Chief Instructor To Be Advised Senior Instructors Wendy Bourke Ron Brouwer Pam Hazelton Peter Stone Donna Brouwer Bonnie Guillan Leslie Rowling Dave Varney Life Members Don Ahearne John Mitchell Bill Barclay* Anne O'Hara * Joan Barclay * Barry Reed Wendy Bourke Noella Smith Ron Brouwer Allan Spratt * Tiki Friezer * Jan Taylor Pam Hazelton Margaret Taylor Margaret Keast Kevin Thomas * Deceased Rene Llewelyn Elizabeth (Betty) Walsh Margaret McGarvey * Peter Waugh Issue SC046-50Y056 March-April 2021 2 Sutherland Shire Dog Training Club Inc Editors Note: Hi Everyone, Welcome to yet another year at SSDTC. And we’re off to a flying start, with a very large enrolment of new members, especially within the first 2 weeks of club . opening. You’re certainly in the right hands with letting us help you train your (Affiliated with the Dogs NSW) dog. -

Table & Ramp Breeds
Judging Operations Department PO Box 900062 Raleigh, NC 27675-9062 919-816-3570 [email protected] www.akc.org TABLE BREEDS SPORTING NON-SPORTING COCKER SPANIEL ALL AMERICAN ESKIMOS ENGLISH COCKER SPANIEL BICHON FRISE NEDERLANDSE KOOIKERHONDJE BOSTON TERRIER COTON DE TULEAR FRENCH BULLDOG HOUNDS LHASA APSO BASENJI LOWCHEN ALL BEAGLES MINIATURE POODLE PETIT BASSET GRIFFON VENDEEN (or Ground) NORWEGIAN LUNDEHUND ALL DACHSHUNDS SCHIPPERKE PORTUGUSE PODENGO PEQUENO SHIBA INU WHIPPET (or Ground or Ramp) TIBETAN SPANIEL TIBETAN TERRIER XOLOITZCUINTLI (Toy and Miniatures) WORKING- NO WORKING BREEDS ON TABLE HERDING CARDIGAN WELSH CORGI TERRIERS MINIATURE AMERICAN SHEPHERD ALL TERRIERS on TABLE, EXCEPT those noted below PEMBROKE WELSH CORGI examined on the GROUND: PULI AIREDALE TERRIER PUMI AMERICAN STAFFORDSHIRE (or Ramp) PYRENEAN SHEPHERD BULL TERRIER SHETLAND SHEEPDOG IRISH TERRIERS (or Ramp) SWEDISH VALLHUND MINI BULL TERRIER (or Table or Ramp) KERRY BLUE TERRIER (or Ramp) FSS/MISCELLANEOUS BREEDS SOFT COATED WHEATEN TERRIER (or Ramp) DANISH-SWEDISH FARMDOG STAFFORDSHIRE BULL TERRIER (or Ramp) LANCASHIRE HEELER MUDI (or Ramp) PERUVIAN INCA ORCHID (Small and Medium) TOY - ALL TOY BREEDS ON TABLE RUSSIAN TOY TEDDY ROOSEVELT TERRIER RAMP OPTIONAL BREEDS At the discretion of the judge through all levels of competition including group and Best in Show judging. AMERICAN WATER SPANIEL STANDARD SCHNAUZERS ENTLEBUCHER MOUNTAIN DOG BOYKIN SPANIEL AMERICAN STAFFORDSHIRE FINNISH LAPPHUND ENGLISH SPRINGER SPANIEL IRISH TERRIERS ICELANDIC SHEEPDOGS FIELD SPANIEL KERRY BLUE TERRIER NORWEGIAN BUHUND LAGOTTO ROMAGNOLO MINI BULL TERRIER (Ground/Table) POLISH LOWLAND SHEEPDOG NS DUCK TOLLING RETRIEVER SOFT COATED WHEATEN TERRIER SPANISH WATER DOG WELSH SPRINGER SPANIEL STAFFORDSHIRE BULL TERRIER MUDI (Misc.) GRAND BASSET GRIFFON VENDEEN FINNISH SPITZ NORRBOTTENSPETS (Misc.) WHIPPET (Ground/Table) BREEDS THAT MUST BE JUDGED ON RAMP Applies to all conformation competition associated with AKC conformation dog shows or at any event at which an AKC conformation title may be earned. -

The History of the BERGER PICARD
The History of the BERGER PICARD By Betsy Richards hought to be the oldest upright ears, resembling in many ways a Beauce, or Beaceron). The mid-length of the French Sheep- Berger Picard of today. coat was ignored for some time, but final- dogs, the Berger Picard Of course it is not certain that the ly recognized as the Berger de Picardie was brought to northern Berger Picard originates strictly from the (or Picard). France and the Pas de Picardie region of France; it is possible, Although the Berger Picard made an Calais during the second even probable, that they were widespread appearance at the first French dog show TCeltic invasion of Gaul around 400 BC. as harsh-coated sheep and cattle dogs in 1863 and were judged in the same class Sheepdogs resembling Berger Picards were typical throughout northwestern as the Beaucerons and Briards, the breed's have been depicted for centuries in tapes- Europe. Some experts insist that this breed rustic appearance did not result in popu- tries, engravings and woodcuts. is related to the more well-known Briard larity as a show dog. In 1898 there was One renowned painting in the Bergerie and Beauceron, while others believe it proof that the Picard was a recognizable Nationale at Rambouillet (the National shares a common origin with Dutch and breed but it was tricolor, piebald with red Sheepfold of France) dating to the start Belgian Shepherds. brown markings. of the 19th century, shows the 1st Master Around the mid-19th century, dogs Picards would continue to be shown Shepherd, Clément Delorme, in the com- used for herding were initially classified and participate in herding trials but strug- pany of a medium-sized, strong-boned dog as one of two types: long hair (Berger de gled for recognition. -

Dog Breeds Pack 1 Professional Vector Graphics Page 1
DOG BREEDS PACK 1 PROFESSIONAL VECTOR GRAPHICS PAGE 1 Affenpinscher Afghan Hound Aidi Airedale Terrier Akbash Akita Inu Alano Español Alaskan Klee Kai Alaskan Malamute Alpine Dachsbracke American American American American Akita American Bulldog Cocker Spaniel Eskimo Dog Foxhound American American Mastiff American Pit American American Hairless Terrier Bull Terrier Staffordshire Terrier Water Spaniel Anatolian Anglo-Français Appenzeller Shepherd Dog de Petite Vénerie Sennenhund Ariege Pointer Ariegeois COPYRIGHT (c) 2013 FOLIEN.DS. ALL RIGHTS RESERVED. WWW.VECTORART.AT DOG BREEDS PACK 1 PROFESSIONAL VECTOR GRAPHICS PAGE 2 Armant Armenian Artois Hound Australian Australian Kelpie Gampr dog Cattle Dog Australian Australian Australian Stumpy Australian Terrier Austrian Black Shepherd Silky Terrier Tail Cattle Dog and Tan Hound Austrian Pinscher Azawakh Bakharwal Dog Barbet Basenji Basque Basset Artésien Basset Bleu Basset Fauve Basset Griffon Shepherd Dog Normand de Gascogne de Bretagne Vendeen, Petit Basset Griffon Bavarian Mountain Vendéen, Grand Basset Hound Hound Beagle Beagle-Harrier COPYRIGHT (c) 2013 FOLIEN.DS. ALL RIGHTS RESERVED. WWW.VECTORART.AT DOG BREEDS PACK 2 PROFESSIONAL VECTOR GRAPHICS PAGE 3 Belgian Shepherd Belgian Shepherd Bearded Collie Beauceron Bedlington Terrier (Tervuren) Dog (Groenendael) Belgian Shepherd Belgian Shepherd Bergamasco Dog (Laekenois) Dog (Malinois) Shepherd Berger Blanc Suisse Berger Picard Bernese Mountain Black and Berner Laufhund Dog Bichon Frisé Billy Tan Coonhound Black and Tan Black Norwegian -

Agtsec Running Groups
Group A - 142 competitors Champ: 10 - 8, 14 - 5, 16 - 12, 20 - 15, 22 - 18, 24 - 12 Perf: P8 - 9, P12 - 6, P14 - 8, P16 - 10, P20 - 9 Vet: V4 - 4, V8 - 5, V12 - 17, V16 - 4 Addison, Michelle DeChance, Annie Lady (24) German Shepherd Dog Pink Floyd (V12) Token Blonde Spencer Davis (P20) Rico Suave Dog Alfonso, Annette Chapter (22) Border Collie Erspamer, Mia Legend (V12) Border Collie Jackson (P20) Labrador Retriever Valid (P20) Border Collie Anderson, Cliff Zoe (20) Wheatable Ferguson, Kelley Winnie (V12) Border Collie Joose (16) Border Collie Anderson, Crystal Floyd, Cindy Razzi (P14) English Springer Spaniel Thor (16) Poodle (Miniature) Andrews, Lisa Friedl, Gwyneth Shibumi (24) Border Collie Amigo (24) Border Collie Aubois, Sara Gant, Shane Ridley (P20) Border Collie Atom (P20) Border Collie Sweets (P12) Shetland Sheepdog Barton, Kim Logan (V 4) Shih Tzu Garcia, Allison EPI (20) All-American Sizzle (V 8) Shetland Sheepdog Better Cheddar (14) All-American Bekaert, Susan Ringer (P16) Border Collie ABBA (V12) Border Collie Motown (16) Shetland Sheepdog Garvey, Sarah Poppy (24) All-American Bennett, Alicia Excalibur (V16) Border Collie Gerhard, Jeremy Bleu (10) Papillon Maverick (10) Pembroke Welsh Corgi Pixie Pig (20) Border Collie Tease (20) Border Collie Ruckus (22) Australian Shepherd Benson, Helen Shadow (16) Shetland Sheepdog Grace, Kathy Blanche (16) Standard Schnauzer Bowman, Tom Casey (P14) All-American Hanson, Morgan Probability (P16) Border Collie Brown, Kat #Winning (22) Border Collie Nemo (P14) All-American Elite (20) Border -

South Australian Obedience Dog Club Inc
South Australian Obedience Dog Club Inc Agility Trials Sunday 9th April 2017 SA Obedience Dog Club Agility Trials Sunday 9th April 2017 SOUTH AUSTRALIAN OBEDIENCE DOG CLUB INC Agility Trials Sunday 9th April 2017 At 9.00 am and not before 1.00 pm 9.00 am Brian Fielder ………………. Agility: Master, Open, Excellent, Novice Jane Lawrence ………….. Jumping: Novice, Excellent, Open, Master Not Before 1.00 pm Rose Ince …………... Agility: Master, Open, Excellent, Novice Julie Lyon………….………Jumping: Novice, Excellent, Open, Master SACA Representative: Mr Victor Jordan Trial Manager: Mrs Barbara Richter-Winter Veterinary: By Committee Vetting 8.00 am to 8.45 am and 12.30 to 12.45 pm. Bitches only without desexing certificate or tattoo. All exhibits to pass checkpoint by close. Order of Judging: Two rings simultaneously as above. One course only for all height categories within each class. Prizes: 1st Place qualifying in each height category Sashes: To all qualifiers SA Obedience Dog Club Agility Trials Sunday 9th April 2017 AM Trial Master Agility – Brian Fielder SCT 60 Time T/F C/F Total Rank 300 Class 3001 Inneslake Royal Kiss CD RN ADM JDM JDO SD GD Sally Millan Shetland Sheepdog D/Q 3004 Rex ADX JDX Rosemary Ince Tenterfield Terrier cross D/Q 3005 Donriver Platinum Edition RN ADM JDM GD S & T Millan Shetland Sheepdog 45.11 0 0 0 Q 1st 3006 Asher CD RE ADX JD GD SD SPDX FS.N HTM.N Julie Brown Papillon cross Sheltie D/Q 3009 Ag Ch (300) Ashem Hera ADM7 JDM5 SDX GDX PT Joan Murray Shetland Sheepdog 59.20 0 0 0 Q 2nd 3010 Gumhaven Totally Cool ADX JDX Andrew -
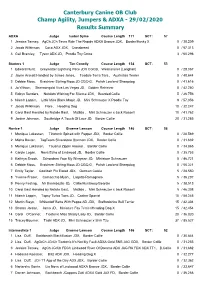
29/02/2020 Results Summary
Canterbury Canine OB Club Champ Agility, Jumpers & ADXA - 29/02/2020 Results Summary ADXA Judge Isobel Syme Course Length 171 SCT: 57 1 Jessica Tansey, AgCh JCh Tevra Ride The Rapids ADXA Bronze JDX, Border/Husky X 0 / 38.209 2 Jacob Wilkinson, Coco ADX JDX, Crossbreed 0 / 57.313 3 Gail Bramley, Tyson ADX JD, Poodle Toy Cross 3 / 60.296 Starters 1 Judge Tim Conolly Course Length 134 SCT: 53 1 Edward Hunt, Greymatter Lightning Pace JDX CGCB, Weimaraner (Longhair) 0 / 28.067 2 Jayne Arscott Handled by James Jones, Tasdale Terra Tara , Australian Terrier 0 / 40.644 3 Debbie Moss, Braidriver Stirling Moss JD CGC-G, Polish Lowland Sheepdog 0 / 41.616 4 Jo Wilson, Shannongold Viva Las Vegas JD, Golden Retriever 0 / 42.250 5 Robyn Sanders, Notslots Wishing For Silence JDX, Bearded Collie 5 / 36.756 6 Niamh Lappin, Little Miss Black Magic JD, Mini Schnauzer X Poodle Toy 9 / 57.356 7 Jacob Wilkinson, Flare , Heading Dog 10 / 32.247 8 Carol East Handled by Natalie East, Matilda , Mini Schnauzer x Jack Russell 10 / 41.762 9 Janine Johnson, Southridge A Touch Of Lace JD, Border Collie 20 / 31.285 Novice 1 Judge Graeme Lawson Course Length 146 SCT: 58 1 Monique Lukassen, Ticotonic Spiced with Pepper JDX, Border Collie 0 / 28.569 2 Misha Baxter, TagTeam Silverstone Starman JDX, Border Collie 0 / 31.632 3 Monique Lukassen, Tauanui Zippin Around , Border Collie 0 / 34.865 4 Carole Logan, Nova Echo of Lindwood JD, Border Collie 0 / 35.753 5 Kathryn Snook, Schondara Your My Wingman JD, Miniature Schnauzer 0 / 46.721 6 Debbie Moss, Braidriver Stirling Moss -

HERDING BREEDS ELIGIBLE to COMPETE in ASCA STOCKDOG TRIALS Ref: the Atlas of Dog Breeds of the World - Bonnie Wilcox, DVM & Chris Walkowic
ASCA STOCKDOG PROGRAM RULES – JUNE 2014 APPENDIX 6: HERDING BREEDS ELIGIBLE TO COMPETE IN ASCA STOCKDOG TRIALS Ref: The Atlas of Dog Breeds of the World - Bonnie Wilcox, DVM & Chris Walkowic Breed Country of Origin Breed Country of Origin Australian Cattle Dog Australia Greater Swiss Mountain Dog Switzerland Australian Kelpie Australia Hairy Mouth Heeler USA Australian Shepherd USA Hovawart Germany Belgian Laekenois Belgium Iceland Dog Iceland Belgian Malinois Belgium Kerry Blue Terrier Ireland Belgian Sheep Dog Lancashire Heeler Great Britain (Groenendael) Belgium Lapinporokoira Belgian Tervuren Belgium (Lapponian Herder) Finland Bouviers Des Flandres Belgium Malinois Belgian Bergamasco Italy McNab USA Bernese Mtn Dog Switzerland Miniature Australian Shepherd USA Beauceron France Mudi Hungary Briard France North American Shepherd USA Bearded Collie Great Britain Norwegian Buhund Norway Border Collie Great Britain Old English Sheep Dog Great Britain Blue Lacy USA Picardy Shepherd France Catahoula Leopard Dog USA Polish Owczarek Nizinny Poland Canaan Dog Israel Puli Hungary Cao de Serra de Aires Pumi Hungary (Portuguese Sheep Dog) Portugal Pyrenean Shepherd France Croatian Sheep Dog Croatia Rottweiler Germany Cataian Sheep Dog Spain Samoyed Scandinavia Collie Great Britain Schapendoes Corgi (Dutch Sheepdog) Netherlands Cardigan Welsh Great Britain Shetland Sheep Dog Great Britain Pembroke Welsh Great Britain Soft Coated Wheaten Terrier Ireland Dutch Shepherd: Netherlands Spanish Water Dog Spain English Shepherd USA Swedish Lapphund Sweden Entlebucher Mountain Dog Switzerland Tibetan Terrier China Finnish Lapphund Finland Vasgotaspets Sweden German Shepherd Dog Germany White Shepherd USA German Coolie/Koolie Australia 74 . -

Australian National Kennel Council Limited National Animal Registration Analysis 2010-2019
AUSTRALIAN NATIONAL KENNEL COUNCIL LIMITED NATIONAL ANIMAL REGISTRATION ANALYSIS 2010-2019 GROUP 1 TOYS 2010 2011 2012 2013 2014 2015 2016 2017 2018 2019 Affenpinscher 17 33 28 51 30 27 25 Australian Silky Terrier 273 277 253 237 199 202 134 Bichon Frise 445 412 445 366 471 465 465 Cavalier King Charles Spaniel 2942 2794 2655 2531 2615 2439 2555 Chihuahua (Long) 690 697 581 565 563 545 520 Chihuahua (Smooth) 778 812 724 713 735 789 771 Chinese Crested Dog 286 248 266 216 221 234 207 Coton De Tulear 0 0 0 0 0 0 2 English Toy Terrier (Black & Tan) 72 61 54 92 58 63 75 Griffon Bruxellois 199 149 166 195 126 166 209 Havanese 158 197 263 320 340 375 415 Italian Greyhound 400 377 342 354 530 521 486 Japanese Chin 66 43 55 61 45 50 53 King Charles Spaniel 29 11 27 29 24 47 25 Lowchen 53 44 67 84 76 59 60 Maltese 315 299 305 296 234 196 185 Miniature Pinscher 209 217 179 227 211 230 195 Papillon 420 415 388 366 386 387 420 Pekingese 186 161 153 172 196 160 173 Pomeranian 561 506 468 410 428 470 471 Pug 1495 1338 1356 1319 1311 1428 1378 Russian Toy 0 0 3 23 37 22 32 Tibetan Spaniel 303 210 261 194 220 226 183 Yorkshire Terrier 237 253 168 234 188 201 227 TOTAL 10134 9554 9207 9055 9244 9302 9266 0 0 0 Page 1 AUSTRALIAN NATIONAL KENNEL COUNCIL LIMITED NATIONAL ANIMAL REGISTRATION ANALYSIS 2010-2019 GROUP 2 TERRIERS 2010 2011 2012 2013 2014 2015 2016 2017 2018 2019 Airedale Terrier 269 203 214 235 308 281 292 American Hairless Terrier 0 0 0 0 0 0 0 American Staffordshire Terrier 1625 2015 1786 2337 2112 2194 1793 Australian Terrier 272 247 308