R-Spondin1/LGR5 Activates Tgfb Signaling and Suppresses Colon
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

KLF2 Induced
UvA-DARE (Digital Academic Repository) The transcription factor KLF2 in vascular biology Boon, R.A. Publication date 2008 Link to publication Citation for published version (APA): Boon, R. A. (2008). The transcription factor KLF2 in vascular biology. General rights It is not permitted to download or to forward/distribute the text or part of it without the consent of the author(s) and/or copyright holder(s), other than for strictly personal, individual use, unless the work is under an open content license (like Creative Commons). Disclaimer/Complaints regulations If you believe that digital publication of certain material infringes any of your rights or (privacy) interests, please let the Library know, stating your reasons. In case of a legitimate complaint, the Library will make the material inaccessible and/or remove it from the website. Please Ask the Library: https://uba.uva.nl/en/contact, or a letter to: Library of the University of Amsterdam, Secretariat, Singel 425, 1012 WP Amsterdam, The Netherlands. You will be contacted as soon as possible. UvA-DARE is a service provided by the library of the University of Amsterdam (https://dare.uva.nl) Download date:23 Sep 2021 Supplementary data: Genes induced by KLF2 Dekker et al. LocusLink Accession Gene Sequence Description Fold p-value ID number symbol change (FDR) 6654 AK022099 SOS1 cDNA FLJ12037 fis, clone HEMBB1001921. 100.00 5.9E-09 56999 AF086069 ADAMTS9 full length insert cDNA clone YZ35C05. 100.00 1.2E-09 6672 AF085934 SP100 full length insert cDNA clone YR57D07. 100.00 6.7E-13 9031 AF132602 BAZ1B Williams Syndrome critical region WS25 mRNA, partial sequence. -

A Computational Approach for Defining a Signature of Β-Cell Golgi Stress in Diabetes Mellitus
Page 1 of 781 Diabetes A Computational Approach for Defining a Signature of β-Cell Golgi Stress in Diabetes Mellitus Robert N. Bone1,6,7, Olufunmilola Oyebamiji2, Sayali Talware2, Sharmila Selvaraj2, Preethi Krishnan3,6, Farooq Syed1,6,7, Huanmei Wu2, Carmella Evans-Molina 1,3,4,5,6,7,8* Departments of 1Pediatrics, 3Medicine, 4Anatomy, Cell Biology & Physiology, 5Biochemistry & Molecular Biology, the 6Center for Diabetes & Metabolic Diseases, and the 7Herman B. Wells Center for Pediatric Research, Indiana University School of Medicine, Indianapolis, IN 46202; 2Department of BioHealth Informatics, Indiana University-Purdue University Indianapolis, Indianapolis, IN, 46202; 8Roudebush VA Medical Center, Indianapolis, IN 46202. *Corresponding Author(s): Carmella Evans-Molina, MD, PhD ([email protected]) Indiana University School of Medicine, 635 Barnhill Drive, MS 2031A, Indianapolis, IN 46202, Telephone: (317) 274-4145, Fax (317) 274-4107 Running Title: Golgi Stress Response in Diabetes Word Count: 4358 Number of Figures: 6 Keywords: Golgi apparatus stress, Islets, β cell, Type 1 diabetes, Type 2 diabetes 1 Diabetes Publish Ahead of Print, published online August 20, 2020 Diabetes Page 2 of 781 ABSTRACT The Golgi apparatus (GA) is an important site of insulin processing and granule maturation, but whether GA organelle dysfunction and GA stress are present in the diabetic β-cell has not been tested. We utilized an informatics-based approach to develop a transcriptional signature of β-cell GA stress using existing RNA sequencing and microarray datasets generated using human islets from donors with diabetes and islets where type 1(T1D) and type 2 diabetes (T2D) had been modeled ex vivo. To narrow our results to GA-specific genes, we applied a filter set of 1,030 genes accepted as GA associated. -

System, Method and Software for Calculation of a Cannabis Drug Efficiency Index for the Reduction of Inflammation
International Journal of Molecular Sciences Article System, Method and Software for Calculation of a Cannabis Drug Efficiency Index for the Reduction of Inflammation Nicolas Borisov 1,† , Yaroslav Ilnytskyy 2,3,†, Boseon Byeon 2,3,4,†, Olga Kovalchuk 2,3 and Igor Kovalchuk 2,3,* 1 Moscow Institute of Physics and Technology, 9 Institutsky lane, Dolgoprudny, Moscow Region 141701, Russia; [email protected] 2 Department of Biological Sciences, University of Lethbridge, Lethbridge, AB T1K 3M4, Canada; [email protected] (Y.I.); [email protected] (B.B.); [email protected] (O.K.) 3 Pathway Rx., 16 Sandstone Rd. S., Lethbridge, AB T1K 7X8, Canada 4 Biomedical and Health Informatics, Computer Science Department, State University of New York, 2 S Clinton St, Syracuse, NY 13202, USA * Correspondence: [email protected] † First three authors contributed equally to this research. Abstract: There are many varieties of Cannabis sativa that differ from each other by composition of cannabinoids, terpenes and other molecules. The medicinal properties of these cultivars are often very different, with some being more efficient than others. This report describes the development of a method and software for the analysis of the efficiency of various cannabis extracts to detect the anti-inflammatory properties of the various cannabis extracts. The method uses high-throughput gene expression profiling data but can potentially use other omics data as well. According to the signaling pathway topology, the gene expression profiles are convoluted into the signaling pathway activities using a signaling pathway impact analysis (SPIA) method. The method was tested by inducing inflammation in human 3D epithelial tissues, including intestine, oral and skin, and then exposing these tissues to various extracts and then performing transcriptome analysis. -

Differential Gene Expression in Oligodendrocyte Progenitor Cells, Oligodendrocytes and Type II Astrocytes
Tohoku J. Exp. Med., 2011,Differential 223, 161-176 Gene Expression in OPCs, Oligodendrocytes and Type II Astrocytes 161 Differential Gene Expression in Oligodendrocyte Progenitor Cells, Oligodendrocytes and Type II Astrocytes Jian-Guo Hu,1,2,* Yan-Xia Wang,3,* Jian-Sheng Zhou,2 Chang-Jie Chen,4 Feng-Chao Wang,1 Xing-Wu Li1 and He-Zuo Lü1,2 1Department of Clinical Laboratory Science, The First Affiliated Hospital of Bengbu Medical College, Bengbu, P.R. China 2Anhui Key Laboratory of Tissue Transplantation, Bengbu Medical College, Bengbu, P.R. China 3Department of Neurobiology, Shanghai Jiaotong University School of Medicine, Shanghai, P.R. China 4Department of Laboratory Medicine, Bengbu Medical College, Bengbu, P.R. China Oligodendrocyte precursor cells (OPCs) are bipotential progenitor cells that can differentiate into myelin-forming oligodendrocytes or functionally undetermined type II astrocytes. Transplantation of OPCs is an attractive therapy for demyelinating diseases. However, due to their bipotential differentiation potential, the majority of OPCs differentiate into astrocytes at transplanted sites. It is therefore important to understand the molecular mechanisms that regulate the transition from OPCs to oligodendrocytes or astrocytes. In this study, we isolated OPCs from the spinal cords of rat embryos (16 days old) and induced them to differentiate into oligodendrocytes or type II astrocytes in the absence or presence of 10% fetal bovine serum, respectively. RNAs were extracted from each cell population and hybridized to GeneChip with 28,700 rat genes. Using the criterion of fold change > 4 in the expression level, we identified 83 genes that were up-regulated and 89 genes that were down-regulated in oligodendrocytes, and 92 genes that were up-regulated and 86 that were down-regulated in type II astrocytes compared with OPCs. -

Comparing Spatial Expression Dynamics of Bovine
Comparing spatial expression dynamics of bovine blastocyst under three different procedures : in-vivo, in-vitro derived, Title and somatic cell nuclear transfer embryos Nagatomo, Hiroaki; Akizawa, Hiroki; Sada, Ayari; Kishi, Yasunori; Yamanaka, Ken-ichi; Takuma, Tetsuya; Sasaki, Author(s) Keisuke; Yamauchi, Nobuhiko; Yanagawa, Yojiro; Nagano, Masashi; Kono, Tomohiro; Takahashi, Masashi; Kawahara, Manabu Citation Japanese Journal of Veterinary Research, 63(4), 159-171 Issue Date 2015-11 DOI 10.14943/jjvr.63.4.159 Doc URL http://hdl.handle.net/2115/60304 Type bulletin (article) Additional Information There are other files related to this item in HUSCAP. Check the above URL. File Information JJVR63-4 p.159-171 Suppl. Table 3.pdf (Supplemental Table 3) Instructions for use Hokkaido University Collection of Scholarly and Academic Papers : HUSCAP Supplemental table 3. Genes that were differentially expressed in the ICM relative to the TE in in SCNT blastocyst (SCNT list). Gene Symbol Probe Set ID Regulation Fold change ([ICM] / [TE]) Gene Title EEF1A1 AFFX-Bt-ef1a-3_at UP 1.2092365 eukaryotic translation elongation factor 1 alpha 1 IGFBP3 Bt.422.1.S2_at UP 2.5323892 insulin-like growth factor binding protein 3 IGFBP3 Bt.422.1.S1_at UP 3.7850845 insulin-like growth factor binding protein 3 SULT1A1 Bt.3537.1.S1_at UP 2.7092714 sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 SPP1 Bt.2632.1.S1_at UP 5.6928325 secreted phosphoprotein 1 SCARB1 Bt.4520.1.S1_at UP 3.106944 scavenger receptor class B, member 1 TSPO Bt.3988.1.S1_at -

Human Induced Pluripotent Stem Cell–Derived Podocytes Mature Into Vascularized Glomeruli Upon Experimental Transplantation
BASIC RESEARCH www.jasn.org Human Induced Pluripotent Stem Cell–Derived Podocytes Mature into Vascularized Glomeruli upon Experimental Transplantation † Sazia Sharmin,* Atsuhiro Taguchi,* Yusuke Kaku,* Yasuhiro Yoshimura,* Tomoko Ohmori,* ‡ † ‡ Tetsushi Sakuma, Masashi Mukoyama, Takashi Yamamoto, Hidetake Kurihara,§ and | Ryuichi Nishinakamura* *Department of Kidney Development, Institute of Molecular Embryology and Genetics, and †Department of Nephrology, Faculty of Life Sciences, Kumamoto University, Kumamoto, Japan; ‡Department of Mathematical and Life Sciences, Graduate School of Science, Hiroshima University, Hiroshima, Japan; §Division of Anatomy, Juntendo University School of Medicine, Tokyo, Japan; and |Japan Science and Technology Agency, CREST, Kumamoto, Japan ABSTRACT Glomerular podocytes express proteins, such as nephrin, that constitute the slit diaphragm, thereby contributing to the filtration process in the kidney. Glomerular development has been analyzed mainly in mice, whereas analysis of human kidney development has been minimal because of limited access to embryonic kidneys. We previously reported the induction of three-dimensional primordial glomeruli from human induced pluripotent stem (iPS) cells. Here, using transcription activator–like effector nuclease-mediated homologous recombination, we generated human iPS cell lines that express green fluorescent protein (GFP) in the NPHS1 locus, which encodes nephrin, and we show that GFP expression facilitated accurate visualization of nephrin-positive podocyte formation in -

Supplemental Table S1. Primers for Sybrgreen Quantitative RT-PCR Assays
Supplemental Table S1. Primers for SYBRGreen quantitative RT-PCR assays. Gene Accession Primer Sequence Length Start Stop Tm GC% GAPDH NM_002046.3 GAPDH F TCCTGTTCGACAGTCAGCCGCA 22 39 60 60.43 59.09 GAPDH R GCGCCCAATACGACCAAATCCGT 23 150 128 60.12 56.52 Exon junction 131/132 (reverse primer) on template NM_002046.3 DNAH6 NM_001370.1 DNAH6 F GGGCCTGGTGCTGCTTTGATGA 22 4690 4711 59.66 59.09% DNAH6 R TAGAGAGCTTTGCCGCTTTGGCG 23 4797 4775 60.06 56.52% Exon junction 4790/4791 (reverse primer) on template NM_001370.1 DNAH7 NM_018897.2 DNAH7 F TGCTGCATGAGCGGGCGATTA 21 9973 9993 59.25 57.14% DNAH7 R AGGAAGCCATGTACAAAGGTTGGCA 25 10073 10049 58.85 48.00% Exon junction 9989/9990 (forward primer) on template NM_018897.2 DNAI1 NM_012144.2 DNAI1 F AACAGATGTGCCTGCAGCTGGG 22 673 694 59.67 59.09 DNAI1 R TCTCGATCCCGGACAGGGTTGT 22 822 801 59.07 59.09 Exon junction 814/815 (reverse primer) on template NM_012144.2 RPGRIP1L NM_015272.2 RPGRIP1L F TCCCAAGGTTTCACAAGAAGGCAGT 25 3118 3142 58.5 48.00% RPGRIP1L R TGCCAAGCTTTGTTCTGCAAGCTGA 25 3238 3214 60.06 48.00% Exon junction 3124/3125 (forward primer) on template NM_015272.2 Supplemental Table S2. Transcripts that differentiate IPF/UIP from controls at 5%FDR Fold- p-value Change Transcript Gene p-value p-value p-value (IPF/UIP (IPF/UIP Cluster ID RefSeq Symbol gene_assignment (Age) (Gender) (Smoking) vs. C) vs. C) NM_001178008 // CBS // cystathionine-beta- 8070632 NM_001178008 CBS synthase // 21q22.3 // 875 /// NM_0000 0.456642 0.314761 0.418564 4.83E-36 -2.23 NM_003013 // SFRP2 // secreted frizzled- 8103254 NM_003013 -

TSPAN2 Sirna (H): Sc-88080
SANTA CRUZ BIOTECHNOLOGY, INC. TSPAN2 siRNA (h): sc-88080 BACKGROUND STORAGE AND RESUSPENSION TSPAN2 (Tetraspanin-2) is a 221 amino acid member of the tetraspanin Store lyophilized siRNA duplex at -20° C with desiccant. Stable for at least (TM4SF) family. Tetraspanins are a group of hydrophobic membrane proteins one year from the date of shipment. Once resuspended, store at -20° C, that interact with a wide variety of proteins including intracellular signaling avoid contact with RNAses and repeated freeze thaw cycles. molecules, integrins and membrane receptors. Members of the tetraspanin Resuspend lyophilized siRNA duplex in 330 µl of the RNAse-free water family are characterized by the presence of four hydrophobic domains and provided. Resuspension of the siRNA duplex in 330 µl of RNAse-free water play a role in cell development, activation, growth and motility. It is believed makes a 10 µM solution in a 10 µM Tris-HCl, pH 8.0, 20 mM NaCl, 1 mM TSPAN2 plays a role in signalling in oligodendrocytes in the early stages of EDTA buffered solution. their terminal differentiation into myelin-forming glia and may also function in stabilizing the mature sheath. TSPAN2 is encoded by a gene located on APPLICATIONS Chromosome 1 which is the largest human chromosome spanning about 260 million base pairs and making up 8% of the human genome. There are about TSPAN2 siRNA (h) is recommended for the inhibition of TSPAN2 expression 3,000 genes on chromosome 1, and considering the great number of genes in human cells. there are also a large number of diseases associated with chromosome 1. -

Tspan2: a Tetraspanin Protein Involved in Oligodendrogenesis and Cancer Metastasis
Biochemical Society Transactions (2017) 45 465–475 DOI: 10.1042/BST20160022 Review Article Tspan2: a tetraspanin protein involved in oligodendrogenesis and cancer metastasis Ibrahim H. Yaseen1, Peter N. Monk2 and Lynda J. Partridge1 1Department of Molecular Biology and Biotechnology, University of Sheffield, Sheffield, U.K. and 2Department of Infection, Immunity and Cardiovascular Diseases, University of Sheffield Medical School, Sheffield, U.K. Correspondence: Ibrahim H. Yaseen ([email protected]) or ([email protected]) Tetraspanin 2 (Tspan2) is one of the less well-characterised members of the tetraspanin superfamily, and its precise function in different human tissue types remains to be explored. Initial studies have highlighted its possible association in neuroinflammation and carcinogenesis. In the central nervous system, Tspan2 may contribute to the early stages of the oligodendrocyte differentiation into myelin-forming glia. Furthermore, in human lung cancer, Tspan2 could be involved in the progression of the tumour metasta- sis by modulating cancer cell motility and invasion functions. In this review, we discuss the available evidence for the potential role of Tspan2 and introduce possible strategies for disease targeting. Introduction Cellular development and pathology are controlled through multiple biological complexes including transmembrane molecules and their associated partners. Tetraspanins, a superfamily of transmem- brane proteins of 20–50 kDa, are abundantly expressed and distributed in multicellular organisms and significantly contribute to health and disease. The tetraspanin family is constituted of 33 members in mammals and retains a highly conserved pattern of amino acid (aa) residues within the family itself, and in evolution with distantly related species [1]. Since the discovery of the family in 1990, research conducted on tetraspanins has shown their wide expression in different tissue and cell types, with most cells expressing several different tetraspanin members [2]. -

TSPAN2 Antibody (N-Term) Affinity Purified Rabbit Polyclonal Antibody (Pab) Catalog # Ap16851a
10320 Camino Santa Fe, Suite G San Diego, CA 92121 Tel: 858.875.1900 Fax: 858.622.0609 TSPAN2 Antibody (N-term) Affinity Purified Rabbit Polyclonal Antibody (Pab) Catalog # AP16851a Specification TSPAN2 Antibody (N-term) - Product Information Application WB,E Primary Accession O60636 Other Accession NP_005716.2 Reactivity Human Host Rabbit Clonality Polyclonal Isotype Rabbit Ig Calculated MW 24148 Antigen Region 25-54 TSPAN2 Antibody (N-term) - Additional Information TSPAN2 Antibody (N-term) (Cat. #AP16851a) Gene ID 10100 western blot analysis in NCI-H292 cell line lysates (35ug/lane).This demonstrates the Other Names Tetraspanin-2, Tspan-2, Tetraspan NET-3, TSPAN2 antibody detected the TSPAN2 TSPAN2 protein (arrow). Target/Specificity This TSPAN2 antibody is generated from TSPAN2 Antibody (N-term) - Background rabbits immunized with a KLH conjugated synthetic peptide between 25-54 amino The protein encoded by this gene is a member acids from the N-terminal region of human of the TSPAN2. transmembrane 4 superfamily, also known as the tetraspanin family. Dilution Most of these members are cell-surface WB~~1:1000 proteins that are characterized by the presence of four Format hydrophobic domains. The Purified polyclonal antibody supplied in PBS proteins mediate signal transduction events with 0.09% (W/V) sodium azide. This that play a role in the antibody is purified through a protein A regulation of cell development, activation, column, followed by peptide affinity growth and motility. purification. TSPAN2 Antibody (N-term) - References Storage Maintain refrigerated at 2-8°C for up to 2 Rose, J.E., et al. Mol. Med. 16 (7-8), 247-253 weeks. -
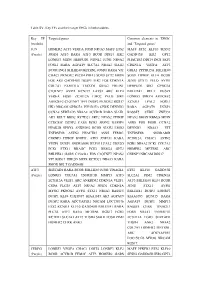
Targeted Genes Common Elements In
Table SV. Key TFs and their target DEGs in hub modules. Key TF Targeted genes Common elements in ‘DEGs’ (module) and ‘Targeted genes’ JUN HOMER2 ATF3 VEGFA FOSB NR4A3 MAFF ETS2 MAFF ETS2 KLF10 SESN2 (Purple) JOSD1 ATF3 RARA ATF3 BCOR DDIT4 IER2 GADD45B IER2 GPT2 LONRF3 MIDN HERPUD1 NDNL2 JUNB NR4A2 PHACTR3 DDIT4 ING1 SKP2 FOSL1 RARA AGPAT9 SLC7A1 NR4A2 SIAH2 CDKN1A VEGFA ATF3 BCOR ING1 BHLHE40 METRNL JOSD1 RARA VIT GRIA2 PPP1R15A BHLHE40 CHAC1 PKNOX2 PLCB4 PHF13 SOX9 SYT2 MIDN SOX9 ITPRIP KLF4 BCOR FOS AK5 GADD45B DUSP1 IER2 FOS CDKN1A JUNB STX11 PELO AVPI1 COL7A1 FAM131A TBCCD1 GRIA2 ERLIN1 HERPUD1 SIK1 GPRC5A C1QTNF7 AVPI1 KCNJ15 LATS2 ARC KLF4 BHLHE41 RELT DUSP2 VEGFA FOSB ZC3H12A LMO2 PELO SIK1 LONRF3 SPRY4 ARHGEF2 ARHGEF2 C1QTNF7 TFPI DUSP2 PKNOX2 RGS17 KCNJ15 LPAL2 FOSL1 HK1 NRCAM GPRC5A PPP1R15A CERK DENND3 RARA AGPAT9 DUSP1 CCNA2 SERTAD3 NR4A2 ACVR1B RARA SIAH1 RASSF5 CERK ZNF331 AK5 RELT BRD2 KCTD21 SKP2 NPAS2 ITPRIP NPAS2 SMOX RBM24 MIDN CCDC85C DUSP2 CARS EGR1 SESN2 RASSF5 ASNS FOS FOSB CCNA2 PIP4K2B SPRY4 ANKRD52 BCOR SIAH2 LMO2 DENND3 NR4A3 VIT TNFRSF1B ASTN2 PHACTR3 ASNS FEM1C TNFRSF1B SNORA80B CSRNP3 ITPRIP BNIP3L ATF3 ZNF331 RARA ZC3H12A CHAC1 ASTN2 VEZF1 DUSP1 SNORA80B KLF10 LPAL2 UBE2O EGR1 NR4A2 PCK1 COL7A1 PCK1 STX11 RRAGC PCK1 RBM24 GPT2 HOMER2 METRNL ARC BHLHE41 GARS C15orf41 FOS C1QTNF7 NPAS2 CSRNP3 NRCAM RGS17 VIT RGS17 UBE2O SOX9 KCTD21 NR4A3 RARA SMOX RELT GADD45B ATF3 SERTAD1 RARA BCOR BHLHE40 JUNB TINAGL1 ETS2 KLF10 GADD45B (Purple) LONRF3 COL7A1 TNFRSF1B MMP13 ATF3 SLC2A1 PIM2 CDKN1A ZC3H12A VEZF1 ARC ANKRD52 -

Fibroblasts from the Human Skin Dermo-Hypodermal Junction Are
cells Article Fibroblasts from the Human Skin Dermo-Hypodermal Junction are Distinct from Dermal Papillary and Reticular Fibroblasts and from Mesenchymal Stem Cells and Exhibit a Specific Molecular Profile Related to Extracellular Matrix Organization and Modeling Valérie Haydont 1,*, Véronique Neiveyans 1, Philippe Perez 1, Élodie Busson 2, 2 1, 3,4,5,6, , Jean-Jacques Lataillade , Daniel Asselineau y and Nicolas O. Fortunel y * 1 Advanced Research, L’Oréal Research and Innovation, 93600 Aulnay-sous-Bois, France; [email protected] (V.N.); [email protected] (P.P.); [email protected] (D.A.) 2 Department of Medical and Surgical Assistance to the Armed Forces, French Forces Biomedical Research Institute (IRBA), 91223 CEDEX Brétigny sur Orge, France; [email protected] (É.B.); [email protected] (J.-J.L.) 3 Laboratoire de Génomique et Radiobiologie de la Kératinopoïèse, Institut de Biologie François Jacob, CEA/DRF/IRCM, 91000 Evry, France 4 INSERM U967, 92260 Fontenay-aux-Roses, France 5 Université Paris-Diderot, 75013 Paris 7, France 6 Université Paris-Saclay, 78140 Paris 11, France * Correspondence: [email protected] (V.H.); [email protected] (N.O.F.); Tel.: +33-1-48-68-96-00 (V.H.); +33-1-60-87-34-92 or +33-1-60-87-34-98 (N.O.F.) These authors contributed equally to the work. y Received: 15 December 2019; Accepted: 24 January 2020; Published: 5 February 2020 Abstract: Human skin dermis contains fibroblast subpopulations in which characterization is crucial due to their roles in extracellular matrix (ECM) biology.