Expression of Cyclomaltodextrinase Gene from Bacillus Halodurans C-125 and Characterization of Its Multisubstrate Specificity
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Bacteria Belonging to Pseudomonas Typographi Sp. Nov. from the Bark Beetle Ips Typographus Have Genomic Potential to Aid in the Host Ecology
insects Article Bacteria Belonging to Pseudomonas typographi sp. nov. from the Bark Beetle Ips typographus Have Genomic Potential to Aid in the Host Ecology Ezequiel Peral-Aranega 1,2 , Zaki Saati-Santamaría 1,2 , Miroslav Kolaˇrik 3,4, Raúl Rivas 1,2,5 and Paula García-Fraile 1,2,4,5,* 1 Microbiology and Genetics Department, University of Salamanca, 37007 Salamanca, Spain; [email protected] (E.P.-A.); [email protected] (Z.S.-S.); [email protected] (R.R.) 2 Spanish-Portuguese Institute for Agricultural Research (CIALE), 37185 Salamanca, Spain 3 Department of Botany, Faculty of Science, Charles University, Benátská 2, 128 01 Prague, Czech Republic; [email protected] 4 Laboratory of Fungal Genetics and Metabolism, Institute of Microbiology of the Academy of Sciences of the Czech Republic, 142 20 Prague, Czech Republic 5 Associated Research Unit of Plant-Microorganism Interaction, University of Salamanca-IRNASA-CSIC, 37008 Salamanca, Spain * Correspondence: [email protected] Received: 4 July 2020; Accepted: 1 September 2020; Published: 3 September 2020 Simple Summary: European Bark Beetle (Ips typographus) is a pest that affects dead and weakened spruce trees. Under certain environmental conditions, it has massive outbreaks, resulting in attacks of healthy trees, becoming a forest pest. It has been proposed that the bark beetle’s microbiome plays a key role in the insect’s ecology, providing nutrients, inhibiting pathogens, and degrading tree defense compounds, among other probable traits. During a study of bacterial associates from I. typographus, we isolated three strains identified as Pseudomonas from different beetle life stages. In this work, we aimed to reveal the taxonomic status of these bacterial strains and to sequence and annotate their genomes to mine possible traits related to a role within the bark beetle holobiont. -

Cyclomaltodextrinase 13A, Paenibacillus Sp. Pcda13a (CBM34-GH13)
Cyclomaltodextrinase 13A, Paenibacillus sp. PCda13A (CBM34-GH13) Catalogue number: CZ08721 , 0,25 mg CZ08722 , 3 × 0,25 mg Description Storage temperature PCda13A (CBM34-GH13), E.C. number 3.2.1.54, is a This enzyme should be stored at -20 °C. cyclomaltodextrinase from Paenibacillus sp. Recombinant PCda13A (CBM34-GH13), purified from Escherichia coli , is a modular family 13 Glycoside Hydrolase (GH13) with an N-terminal Substrate specificity family 34 Carbohydrate Binding Module (CBM34) (www.cazy.org). The enzyme is provided in 35 mM NaHepes buffer, pH 7.5, 750 PCda13A (CBM34-GH13) hydrolyses α-, β-, and γ-cyclodextrins (CDs). The enzyme hydrolyze CDs and linear maltooligosaccharides mM NaCl, 200 mM imidazol, 3.5 mM CaCl 2 and 25% (v/v) glycerol, at a 0,25 mg/mL concentration. Bulk quantities of this product are to yield maltose and glucose with less amounts of maltotriose and available on request. maltotetraose. Electrophoretic Purity Temperature and pH optima PCda13A (CBM34-GH13) purity was determined by sodium The pH optimum for enzymatic activity is 7 while temperature dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) optimum is 40 °C. followed by BlueSafe staining (MB15201) (Figure 1). Enzyme activity Substrate specificity and kinetic properties of PCda13A (CBM34- GH13) are described in the reference provided below. Follow the instructions described in the paper for the implementation of enzyme assays and to obtain values of specific activity. To measure catalytic activity of GHs, quantify reducing sugars released from polysaccharides through the method described by Miller (1959; Anal. Chem. 31, 426-428). Reference Kaulpiboon and Pongsawasdi. J Biochem Mol Biol. -

Proteome Cold-Shock Response in the Extremely Acidophilic Archaeon, Cuniculiplasma Divulgatum
microorganisms Article Proteome Cold-Shock Response in the Extremely Acidophilic Archaeon, Cuniculiplasma divulgatum Rafael Bargiela 1 , Karin Lanthaler 1,2, Colin M. Potter 1,2 , Manuel Ferrer 3 , Alexander F. Yakunin 1,2, Bela Paizs 1,2, Peter N. Golyshin 1,2 and Olga V. Golyshina 1,2,* 1 School of Natural Sciences, Bangor University, Deiniol Rd, Bangor LL57 2UW, UK; [email protected] (R.B.); [email protected] (K.L.); [email protected] (C.M.P.); [email protected] (A.F.Y.); [email protected] (B.P.); [email protected] (P.N.G.) 2 Centre for Environmental Biotechnology, Bangor University, Deiniol Rd, Bangor LL57 2UW, UK 3 Systems Biotechnology Group, Department of Applied Biocatalysis, CSIC—Institute of Catalysis, Marie Curie 2, 28049 Madrid, Spain; [email protected] * Correspondence: [email protected]; Tel.: +44-1248-388607; Fax: +44-1248-382569 Received: 27 April 2020; Accepted: 15 May 2020; Published: 19 May 2020 Abstract: The archaeon Cuniculiplasma divulgatum is ubiquitous in acidic environments with low-to-moderate temperatures. However, molecular mechanisms underlying its ability to thrive at lower temperatures remain unexplored. Using mass spectrometry (MS)-based proteomics, we analysed the effect of short-term (3 h) exposure to cold. The C. divulgatum genome encodes 2016 protein-coding genes, from which 819 proteins were identified in the cells grown under optimal conditions. In line with the peptidolytic lifestyle of C. divulgatum, its intracellular proteome revealed the abundance of proteases, ABC transporters and cytochrome C oxidase. From 747 quantifiable polypeptides, the levels of 582 proteins showed no change after the cold shock, whereas 104 proteins were upregulated suggesting that they might be contributing to cold adaptation. -

Function and Structure Studies of GH Family 31 and 97 \Alpha
110610 (RV-17) Biosci. Biotechnol. Biochem., 75 (12), 110610-1–9, 2011 Award Review Function and Structure Studies of GH Family 31 and 97 -Glycosidases Masayuki OKUYAMA Research Faculty of Agriculture, Hokkaido University, Kita-9, Nishi-9, Kita-ku, Sapporo 060-8589, Japan Online Publication, December 7, 2011 [doi:10.1271/bbb.110610] A huge number of glycoside hydrolases are classified an evolutionary relationship of proteins in the family, into the glycoside hydrolase family (GH family) based from which it is often possible to extract information on on their amino-acid sequence similarity. The glycoside function and structure.3) Classification in a GH family hydrolases acting on -glucosidic linkage are in GH has, accordingly, become indispensable for research on family 4, 13, 15, 31, 63, 97, and 122. This review deals glycoside hydrolases. Glycoside hydrolases are also mainly with findings on GH family 31 and 97 enzymes. divided into two mechanistic classes, inverting and Research on two GH family 31 enzymes is described: retaining enzymes: with inversion or net retention of clarification of the substrate recognition of Escherichia anomeric configuration during the catalytic reaction.4) coli -xylosidase, and glycosynthase derived from Schiz- The stereochemical outcome is generally conserved in a osaccharomyces pombe -glucosidase. GH family 97 is GH family. The inverting mechanism proceeds via a an aberrant GH family, containing inverting and simple single displacement. Two functional groups, retainingAdvance glycoside hydrolases. The inverting View enzyme usually carboxyl groups, act as general acid and general in GH family 97 displays significant similarity to base catalysts. The general acid catalyst donates a proton retaining -glycosidases, including GH family 97 retain- to the departure aglycon, and the general base catalyst ing -glycosidase, but the inverting enzyme has no simultaneously deprotonates the incoming water mole- catalytic nucleophile residue. -

A Korarchaeal Genome Reveals Insights Into the Evolution of the Archaea
A korarchaeal genome reveals insights into the evolution of the Archaea James G. Elkinsa,b, Mircea Podarc, David E. Grahamd, Kira S. Makarovae, Yuri Wolfe, Lennart Randauf, Brian P. Hedlundg, Ce´ line Brochier-Armaneth, Victor Kunini, Iain Andersoni, Alla Lapidusi, Eugene Goltsmani, Kerrie Barryi, Eugene V. Koonine, Phil Hugenholtzi, Nikos Kyrpidesi, Gerhard Wannerj, Paul Richardsoni, Martin Kellerc, and Karl O. Stettera,k,l aLehrstuhl fu¨r Mikrobiologie und Archaeenzentrum, Universita¨t Regensburg, D-93053 Regensburg, Germany; cBiosciences Division, Oak Ridge National Laboratory, Oak Ridge, TN 37831; dDepartment of Chemistry and Biochemistry, University of Texas, Austin, TX 78712; eNational Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bethesda, MD 20894; fDepartment of Molecular Biophysics and Biochemistry, Yale University, New Haven, CT 06520; gSchool of Life Sciences, University of Nevada, Las Vegas, NV 89154; hLaboratoire de Chimie Bacte´rienne, Unite´ Propre de Recherche 9043, Centre National de la Recherche Scientifique, Universite´de Provence Aix-Marseille I, 13331 Marseille Cedex 3, France; iU.S. Department of Energy Joint Genome Institute, Walnut Creek, CA 94598; jInstitute of Botany, Ludwig Maximilians University of Munich, D-80638 Munich, Germany; and kInstitute of Geophysics and Planetary Physics, University of California, Los Angeles, CA 90095 Communicated by Carl R. Woese, University of Illinois at Urbana–Champaign, Urbana, IL, April 2, 2008 (received for review January 7, 2008) The candidate division Korarchaeota comprises a group of uncul- and sediment samples from Obsidian Pool as an inoculum. The tivated microorganisms that, by their small subunit rRNA phylog- cultivation system supported the stable growth of a mixed commu- eny, may have diverged early from the major archaeal phyla nity of hyperthermophilic bacteria and archaea including an or- Crenarchaeota and Euryarchaeota. -

Archaeal Adaptation to Higher Temperatures Revealed by Genomic Sequence of Thermoplasma Volcanium
Archaeal adaptation to higher temperatures revealed by genomic sequence of Thermoplasma volcanium Tsuyoshi Kawashima*†, Naoki Amano*†‡, Hideaki Koike*†, Shin-ichi Makino†, Sadaharu Higuchi†, Yoshie Kawashima-Ohya†, Koji Watanabe§, Masaaki Yamazaki§, Keiichi Kanehori¶, Takeshi Kawamotoʈ, Tatsuo Nunoshiba**, Yoshihiro Yamamoto††, Hironori Aramaki‡‡, Kozo Makino§§, and Masashi Suzuki†¶¶ †National Institute of Bioscience and Human Technology, Core Research for Evolutional Science and Technology Centre of Structural Biology, 1-1 Higashi, Tsukuba 305-0046, Japan; ‡Doctoral Program in Medical Sciences, University of Tsukuba, 1-1-1 Tennohdai, Tsukuba 305-0006, Japan; §Bioscience Research Laboratory, Fujiya, 228 Soya, Hadano 257-0031, Japan; ¶DNA Analysis Department, Techno Research Laboratory, Hitachi Science Systems, 1-280 Higashi-Koigakubo, Kokubunji 185-8601, Japan; ʈDepartment of Biochemistry, Hiroshima University, School of Dentistry, 1-2-3 Kasumi, Minami-ku, Hiroshima 734-8553, Japan; **Department of Molecular and Cellular Biology, Biological Institute, Graduate School of Science, Tohoku University, Sendai 980-8578, Japan; ††Department of Genetics, Hyogo College of Medicine, Nishinomiya 663-8501, Japan; ‡‡Department of Molecular Biology, Daiichi College of Pharmaceutical Science, 22-1 Tamagawa-cho, Minami-ku, Fukuoka 815-8511, Japan; and §§Department of Molecular Microbiology, The Research Institute of Microbial Diseases, Osaka University, 3-1 Yamadaoka, Suita 565-0871, Japan Edited by Aaron Klug, Royal Society of London, London, United Kingdom, and approved October 16, 2000 (received for review August 18, 2000) The complete genomic sequence of the archaeon Thermoplasma contigs. The remaining gaps were bridged by DNA fragments volcanium, possessing optimum growth temperature (OGT) of constructed using the PCR. The average repetition in sequencing 60°C, is reported. By systematically comparing this genomic se- the same base positions was 13-fold. -

The Main (Glyco) Phospholipid (MPL) of Thermoplasma Acidophilum
International Journal of Molecular Sciences Review The Main (Glyco) Phospholipid (MPL) of Thermoplasma acidophilum Hans-Joachim Freisleben 1,2 1 Goethe-Universität, Gustav-Embden-Zentrum, Laboratory of Microbiological Chemistry, Theodor-Stern-Kai 7, D-60590 Frankfurt am Main, Germany; [email protected] 2 Universitas Indonesia, Medical Research Unit, Faculty of Medicine, Jalan Salemba Raya 6, Jakarta 10430, Indonesia Received: 19 September 2019; Accepted: 18 October 2019; Published: 21 October 2019 Abstract: The main phospholipid (MPL) of Thermoplasma acidophilum DSM 1728 was isolated, purified and physico-chemically characterized by differential scanning calorimetry (DSC)/differential thermal analysis (DTA) for its thermotropic behavior, alone and in mixtures with other lipids, cholesterol, hydrophobic peptides and pore-forming ionophores. Model membranes from MPL were investigated; black lipid membrane, Langmuir-Blodgett monolayer, and liposomes. Laboratory results were compared to computer simulation. MPL forms stable and resistant liposomes with highly proton-impermeable membrane and mixes at certain degree with common bilayer-forming lipids. Monomeric bacteriorhodopsin and ATP synthase from Micrococcus luteus were co-reconstituted and light-driven ATP synthesis measured. This review reports about almost four decades of research on Thermoplasma membrane and its MPL as well as transfer of this research to Thermoplasma species recently isolated from Indonesian volcanoes. Keywords: Thermoplasma acidophilum; Thermoplasma volcanium; -

(Agus Kurnia)Ok
HAYATI Journal of Biosciences September 2012 Available online at: Vol. 19 No. 3, p 150-154 http://journal.ipb.ac.id/index.php/hayati EISSN: 2086-4094 DOI: 10.4308/hjb.19.3.150 SHORT COMMUNICATION Archaeal Life on Tangkuban Perahu- Sampling and Culture Growth in Indonesian Laboratories SRI HANDAYANI1, IMAN SANTOSO1, HANS-JOACHIM FREISLEBEN2∗, HARALD HUBER3, ANDI1, FERY ARDIANSYAH1, CENMI MULYANTO1, ZESSINDA LUTHFA1, ROSARI SALEH1, SERUNI KUSUMA UDYANINGSIH FREISLEBEN1, SEPTELIA INAWATI WANANDI2, MICHAEL THOMM3 1Faculty of Mathematics and Natural Sciences, Universitas Indonesia, Jakarta-Depok, Jakarta 10430, Indonesia 2Faculty of Medicine, Universitas Indonesia, Jalan Salemba Raya No. 6, Jakarta 10430, Indonesia 3Department of Microbiology, Archaea Centre, University of Regensburg, Germany Received May 9, 2012/Accepted September 21, 2012 The aim of the expedition to Tangkuban Perahu, West Java was to obtain archaeal samples from the solfatara fields located in Domas crater. This was one of the places, where scientists from the University of Regensburg Germany had formerly isolated Indonesian archaea, especially Thermoplasma and Sulfolobus species but not fully characterized. We collected five samples from mud holes with temperatures from 57 to 88 oC and pH of 1.5-2. A portion of each sample was grown at the University of Regensburg in modified Allen’s medium at 80 oC. From four out of five samples enrichment cultures were obtained, autotrophically on elemental sulphur and heterotrophically on sulfur and yeast extract; electron micrographs are presented. In the laboratories of Universitas Indonesia the isolates were cultured at 55-60 oC in order to grow tetraetherlipid synthesizing archaea, both Thermoplasmatales and Sulfolobales. Here, we succeeded to culture the same type of archaeal cells, which had been cultured in Regensburg, probably a Sulfolobus species and in Freundt’s medium, Thermoplasma species. -

Characterization of Starch Debranching Enzymes of Maize Endosperm Afroza Rahman Iowa State University
Iowa State University Capstones, Theses and Retrospective Theses and Dissertations Dissertations 1998 Characterization of starch debranching enzymes of maize endosperm Afroza Rahman Iowa State University Follow this and additional works at: https://lib.dr.iastate.edu/rtd Part of the Biochemistry Commons, Molecular Biology Commons, and the Plant Sciences Commons Recommended Citation Rahman, Afroza, "Characterization of starch debranching enzymes of maize endosperm " (1998). Retrospective Theses and Dissertations. 12519. https://lib.dr.iastate.edu/rtd/12519 This Dissertation is brought to you for free and open access by the Iowa State University Capstones, Theses and Dissertations at Iowa State University Digital Repository. It has been accepted for inclusion in Retrospective Theses and Dissertations by an authorized administrator of Iowa State University Digital Repository. For more information, please contact [email protected]. INFORMATION TO USERS This manuscript has been reproduced from the microfilm master. UMI films the text directly from the original or copy submitted. Thus, some thesis and dissertation copies are in typewriter face, while others may be from any type of computer printer. The quality of this reproduction is dependent upon the quality of the copy submitted. Broken or indistinct print, colored or poor quality illustrations and photographs, print bleedthrough, substandard margins, and improper alignment can adversely affect reproduction. In the unlikely event that the author did not send UMI a complete manuscript and there are missing pages, these will be noted. Also, if unauthorized copyright material had to be removed, a note will indicate the deletion. Oversize materials (e.g., maps, drawings, charts) are reproduced by sectioning the original, beginning at the upper left-hand comer and continuing from left to right in equal sections with small overlaps. -

(12) United States Patent (10) Patent No.: US 9,689,046 B2 Mayall Et Al
USOO9689046B2 (12) United States Patent (10) Patent No.: US 9,689,046 B2 Mayall et al. (45) Date of Patent: Jun. 27, 2017 (54) SYSTEM AND METHODS FOR THE FOREIGN PATENT DOCUMENTS DETECTION OF MULTIPLE CHEMICAL WO O125472 A1 4/2001 COMPOUNDS WO O169245 A2 9, 2001 (71) Applicants: Robert Matthew Mayall, Calgary (CA); Emily Candice Hicks, Calgary OTHER PUBLICATIONS (CA); Margaret Mary-Flora Bebeselea, A. et al., “Electrochemical Degradation and Determina Renaud-Young, Calgary (CA); David tion of 4-Nitrophenol Using Multiple Pulsed Amperometry at Christopher Lloyd, Calgary (CA); Lisa Graphite Based Electrodes', Chem. Bull. “Politehnica” Univ. Kara Oberding, Calgary (CA); Iain (Timisoara), vol. 53(67), 1-2, 2008. Fraser Scotney George, Calgary (CA) Ben-Yoav. H. et al., “A whole cell electrochemical biosensor for water genotoxicity bio-detection”. Electrochimica Acta, 2009, 54(25), 6113-6118. (72) Inventors: Robert Matthew Mayall, Calgary Biran, I. et al., “On-line monitoring of gene expression'. Microbi (CA); Emily Candice Hicks, Calgary ology (Reading, England), 1999, 145 (Pt 8), 2129-2133. (CA); Margaret Mary-Flora Da Silva, P.S. et al., “Electrochemical Behavior of Hydroquinone Renaud-Young, Calgary (CA); David and Catechol at a Silsesquioxane-Modified Carbon Paste Elec trode'. J. Braz. Chem. Soc., vol. 24, No. 4, 695-699, 2013. Christopher Lloyd, Calgary (CA); Lisa Enache, T. A. & Oliveira-Brett, A. M., "Phenol and Para-Substituted Kara Oberding, Calgary (CA); Iain Phenols Electrochemical Oxidation Pathways”, Journal of Fraser Scotney George, Calgary (CA) Electroanalytical Chemistry, 2011, 1-35. Etesami, M. et al., “Electrooxidation of hydroquinone on simply prepared Au-Pt bimetallic nanoparticles'. Science China, Chem (73) Assignee: FREDSENSE TECHNOLOGIES istry, vol. -
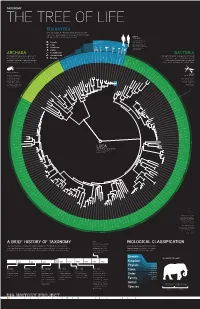
A Brief History of Taxonomy Biological Classification
TAXONOMY THE TREE OF LIFE EUKARYOTA This domain includes all of the plants, animals, and fungi, and some single-celled organisms. Eukaryotes are distinguished by their complex cells, which contain a membrane-enclosed nucleus. Humans Homo sapiens The creatures most familiar to us, Our species, primates in the animals, are members of the Animalia kingdom of the Animalia same kingdom. Eukaryota, is thought to have Fungi Mosquito Red first evolved in Africa about Pufferfish Junglefowl Roundworm Mouse 200,000 years ago. Genetically, Amoebozoa Chimpanzee our closest living relative Plantae is the chimpanzee. Archaeplastida Schizosaccharomyces pombe ARCHAEA Saccharomyces cerevisiae BACTERIA Caenorhabditis briggsae Caenorhabditis elegans Eremothecium gossypii These single-celled prokaryotic organisms often Chromalveolata Dictyostelium discoideum These single-celled prokaryotic organisms were among Cyanidioschyzon merolae live in extreme environmental conditions. Once Excavata Arabidopsis thaliana the first life forms to appear on Earth. Often spherical, Plasmodium falciparum considered to be Bacteria, these microorganisms Cryptosporidium hominis rod-like, or spiral in shape, these microorganisms Thalassiosira pseudonana Oryza sativa Anopheles gambiae Drosophila melanogaster Takifugu rubripes Danio rerio are now recognized as a separate domain of life. Gallus gallus function without a membrane-enclosed cell nucleus. Rattus norvegicus Mus musculus Methanococcus jannaschii Leishmania major Homo sapiens Pan troglodytes Methanococcus maripaludi Thermoanaerobacter -

Temperature and Elemental Sulfur Shape Microbial Communities in Two Extremely Acidic Aquatic Volcanic Environments
bioRxiv preprint doi: https://doi.org/10.1101/2020.09.24.312660; this version posted September 25, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. Temperature and elemental sulfur shape microbial communities in two extremely acidic aquatic volcanic environments Diego Rojas-Gätjens1, Alejandro Arce-Rodríguez2α, Fernando Puente-Sánchez3, Roberto Avendaño1, Eduardo Libby4, Raúl Mora-Amador5,6, Keilor Rojas-Jimenez7, Paola Fuentes-Schweizer4,8, Dietmar H. Pieper2 & Max Chavarría1,4,9* 1Centro Nacional de Innovaciones Biotecnológicas (CENIBiot), CeNAT-CONARE, 1174-1200, San José, Costa Rica 2Microbial Interactions and Processes Research Group, Helmholtz Centre for Infection Research, 38124, Braunschweig, Germany 3Systems Biology Program, Centro Nacional de Biotecnología (CNB-CSIC), C/Darwin 3, 28049 Madrid, Spain 4Escuela de Química, Universidad de Costa Rica, 11501-2060, San José, Costa Rica 5Escuela Centroamericana de Geología, Universidad de Costa Rica, 11501-2060, San José, Costa Rica 6Laboratorio de Ecología Urbana, Universidad Estatal a Distancia, 11501-2060, San José, Costa Rica 7Escuela de Biología, Universidad de Costa Rica, 11501-2060, San José, Costa Rica, 8Centro de Investigación en Electroquímica y Energía Química (CELEQ), Universidad de Costa Rica, 11501-2060, San José, Costa Rica 9Centro de Investigaciones en Productos Naturales