Identification/Selection of E-CAM MD Codes for Development
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

MODELLER 10.1 Manual
MODELLER A Program for Protein Structure Modeling Release 10.1, r12156 Andrej Saliˇ with help from Ben Webb, M.S. Madhusudhan, Min-Yi Shen, Guangqiang Dong, Marc A. Martı-Renom, Narayanan Eswar, Frank Alber, Maya Topf, Baldomero Oliva, Andr´as Fiser, Roberto S´anchez, Bozidar Yerkovich, Azat Badretdinov, Francisco Melo, John P. Overington, and Eric Feyfant email: modeller-care AT salilab.org URL https://salilab.org/modeller/ 2021/03/12 ii Contents Copyright notice xxi Acknowledgments xxv 1 Introduction 1 1.1 What is Modeller?............................................. 1 1.2 Modeller bibliography....................................... .... 2 1.3 Obtainingandinstallingtheprogram. .................... 3 1.4 Bugreports...................................... ............ 4 1.5 Method for comparative protein structure modeling by Modeller ................... 5 1.6 Using Modeller forcomparativemodeling. ... 8 1.6.1 Preparinginputfiles . ............. 8 1.6.2 Running Modeller ......................................... 9 2 Automated comparative modeling with AutoModel 11 2.1 Simpleusage ..................................... ............ 11 2.2 Moreadvancedusage............................... .............. 12 2.2.1 Including water molecules, HETATM residues, and hydrogenatoms .............. 12 2.2.2 Changing the default optimization and refinement protocol ................... 14 2.2.3 Getting a very fast and approximate model . ................. 14 2.2.4 Building a model from multiple templates . .................. 15 2.2.5 Buildinganallhydrogenmodel -

MODELLER 10.0 Manual
MODELLER A Program for Protein Structure Modeling Release 10.0, r12005 Andrej Saliˇ with help from Ben Webb, M.S. Madhusudhan, Min-Yi Shen, Guangqiang Dong, Marc A. Martı-Renom, Narayanan Eswar, Frank Alber, Maya Topf, Baldomero Oliva, Andr´as Fiser, Roberto S´anchez, Bozidar Yerkovich, Azat Badretdinov, Francisco Melo, John P. Overington, and Eric Feyfant email: modeller-care AT salilab.org URL https://salilab.org/modeller/ 2021/01/25 ii Contents Copyright notice xxi Acknowledgments xxv 1 Introduction 1 1.1 What is Modeller?............................................. 1 1.2 Modeller bibliography....................................... .... 2 1.3 Obtainingandinstallingtheprogram. .................... 3 1.4 Bugreports...................................... ............ 4 1.5 Method for comparative protein structure modeling by Modeller ................... 5 1.6 Using Modeller forcomparativemodeling. ... 8 1.6.1 Preparinginputfiles . ............. 8 1.6.2 Running Modeller ......................................... 9 2 Automated comparative modeling with AutoModel 11 2.1 Simpleusage ..................................... ............ 11 2.2 Moreadvancedusage............................... .............. 12 2.2.1 Including water molecules, HETATM residues, and hydrogenatoms .............. 12 2.2.2 Changing the default optimization and refinement protocol ................... 14 2.2.3 Getting a very fast and approximate model . ................. 14 2.2.4 Building a model from multiple templates . .................. 15 2.2.5 Buildinganallhydrogenmodel -

JRC QSAR Model Database
JRC QSAR Model Database EURL ECVAM DataBase service on ALternative Methods to animal experimentation To promote the development and uptake of alternative and advanced methods in toxicology and biomedical sciences SDF - STRUCTURE DATA FORMAT: How to create from SMILES The European Commission’s science and knowledge service Joint Research Centre Directorate F Health, Consumers & Reference Materials Chemicals Safety & Alternative Methods Unit The European Commission’s science and knowledge service Joint Research Centre EUR 28708 EN This publication is a Tutorial by the Joint Research Centre (JRC), the European Commission’s science and knowledge service. It aims to provide user support. The scientific output expressed does not imply a policy position of the European Commission. Neither the European Commission nor any person acting on behalf of the Commission is responsible for the use that might be made of this publication. Contact information Email: [email protected] JRC Science Hub https://ec.europa.eu/jrc JRC107492 EUR 28708 EN PDF ISBN 978-92-79-71294-4 ISSN 1831-9424 doi:10.2760/952280 Print ISBN 978-92-79-71295-1 ISSN 1018-5593 doi:10.2760/668595 Luxembourg: Publications Office of the European Union, 2017 Ispra: European Commission, 2017 © European Union, 2017 The reuse of the document is authorised, provided the source is acknowledged and the original meaning or message of the texts are not distorted. The European Commission shall not be held liable for any consequences stemming from the reuse. How to cite this document: Triebe -

Molecular Docking Studies of a Cyclic Octapeptide-Cyclosaplin from Sandalwood
Preprints (www.preprints.org) | NOT PEER-REVIEWED | Posted: 11 June 2019 doi:10.20944/preprints201906.0091.v1 Peer-reviewed version available at Biomolecules 2019, 9; doi:10.3390/biom9040123 Molecular Docking Studies of a Cyclic Octapeptide-Cyclosaplin from Sandalwood Abheepsa Mishra1, 2,* and Satyahari Dey1 1Plant Biotechnology Laboratory, Department of Biotechnology, Indian Institute of Technology Kharagpur, Kharagpur-721302, West Bengal, India; [email protected] (A.M.); [email protected] (S.D.) 2Department of Internal Medicine, The University of Texas Southwestern Medical Center, 5323 Harry Hines Blvd, Dallas, TX 75390, USA *Correspondence: [email protected]; Tel.:+1(518) 881-9196 Abstract Natural products from plants such as, chemopreventive agents attract huge attention because of their low toxicity and high specificity. The rational drug design in combination with structure based modeling and rapid screening methods offer significant potential for identifying and developing lead anticancer molecules. Thus, the molecular docking method plays an important role in screening a large set of molecules based on their free binding energies and proposes structural hypotheses of how the molecules can inhibit the target. Several peptide based therapeutics have been developed to combat several health disorders including cancers, metabolic disorders, heart-related, and infectious diseases. Despite the discovery of hundreds of such therapeutic peptides however, only few peptide-based drugs have made it to the market. Moreover, until date the activities of cyclic peptides towards molecular targets such as protein kinases, proteases, and apoptosis related proteins have never been explored. In this study we explore the in silico kinase and protease inhibitor potentials of cyclosaplin as well as study the interactions of cyclosaplin with other cancer-related proteins. -
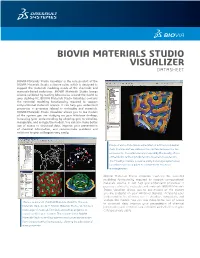
Biovia Materials Studio Visualizer Datasheet
BIOVIA MATERIALS STUDIO VISUALIZER DATASHEET BIOVIA Materials Studio Visualizer is the core product of the BIOVIA Materials Studio software suite, which is designed to support the materials modeling needs of the chemicals and materials-based industries. BIOVIA Materials Studio brings science validated by leading laboratories around the world to your desktop PC. BIOVIA Materials Studio Visualizer contains the essential modeling functionality required to support computational materials science. It can help you understand properties or processes related to molecules and materials. BIOVIA Materials Studio Visualizer allows you to see models of the system you are studying on your Windows desktop, increasing your understanding by allowing you to visualize, manipulate, and analyze the models. You can also make better use of access to structural data, improve your presentation of chemical information, and communicate problems and solutions to your colleagues very easily. Image of early-stage phase segregation in a diblock copolymer melt. The blue surface indicates the interface between the two components. The volume is colormapped by the density of one of the blocks, red being high density, blue being low-density. The MesoDyn module is used to study these large systems over long-times such as required to observe these structural rearrangements. BIOVIA Materials Studio Visualizer contains the essential modeling functionality required to support computational materials science. It can help you understand properties or processes related to molecules and materials. BIOVIA Materials Studio Visualizer allows you to see models of the system you are studying on your Windows desktop, increasing your understanding by allowing you to visualize, manipulate, and analyze the models. -

BIOVIA DISCOVERY STUDIO® 2016 COMPREHENSIVE MODELING and SIMULATIONS for LIFE SCIENCES Datasheet
BIOVIA DISCOVERY STUDIO® 2016 COMPREHENSIVE MODELING AND SIMULATIONS FOR LIFE SCIENCES Datasheet ACCURATELY Drug discovery is a multi-objective optimization. Scientists have to optimize both biochemical potency and characteristics such as ADME and toxicity. The latest PREDICT LIGAND release of BIOVIA’s predictive science application, Discovery Studio®, continues the BINDING evolution of new science in its market-leading CHARMm molecular simulations engine. Built on BIOVIA Foundation™, Discovery Studio® is uniquely positioned as ENERGIES the most comprehensive, collaborative modeling and simulation application for Life Sciences discovery research. DISCOVERY STUDIO 2016 Part of the 2016 BIOVIA product release series, Discovery Studio 2016 continues to deliver key new CHARMm-based molecular simulations. NEW AND ENHANCED SCIENCE • New! Steered Molecular Dynamics: Developed and validated in academia by members of the CHARMM Developer community2,3, the CHARMM AFM (Atomic Force Microscopy) function has been included in the latest release of Discovery Studio CHARMm • Apply a pull force to a molecular system to: • Estimate the ligand binding free energy • Study the conformational details of the ligand unbinding process • Investigate protein unfolding or conformational • Major DMol3 Performance Improvement: The latest release changes of the density functional theory program DMol3, version • Two protocols have been included to enable the simulation 2016, includes dramatic performance improvements, both in of protein or protein-ligand complexes while -

In Silico Screening and Molecular Docking of Bioactive Agents Towards Human Coronavirus Receptor
GSC Biological and Pharmaceutical Sciences, 2020, 11(01), 132–140 Available online at GSC Online Press Directory GSC Biological and Pharmaceutical Sciences e-ISSN: 2581-3250, CODEN (USA): GBPSC2 Journal homepage: https://www.gsconlinepress.com/journals/gscbps (RESEARCH ARTICLE) In silico screening and molecular docking of bioactive agents towards human coronavirus receptor Pratyush Kumar *, Asnani Alpana, Chaple Dinesh and Bais Abhinav Priyadarshini J. L. College of Pharmacy, Electronic Building, Electronic Zone, MIDC, Hingna Road, Nagpur-440016, Maharashtra, India. Publication history: Received on 09 April 2020; revised on 13 April 2020; accepted on 15 April 2020 Article DOI: https://doi.org/10.30574/gscbps.2020.11.1.0099 Abstract Coronavirus infection has turned into pandemic despite of efforts of efforts of countries like America, Italy, China, France etc. Currently India is also outraged by the virulent effect of coronavirus. Although World Health Organisation initially claimed to have all controls over the virus, till date infection has coasted several lives worldwide. Currently we do not have enough time for carrying out traditional approaches of drug discovery. Computer aided drug designing approaches are the best solution. The present study is completely dedicated to in silico approaches like virtual screening, molecular docking and molecular property calculation. The library of 15 bioactive molecules was built and virtual screening was carried towards the crystalline structure of human coronavirus (6nzk) which was downloaded from protein database. Pyrx virtual screening tool was used and results revealed that F14 showed best binding affinity. The best screened molecule was further allowed to dock with the target using Autodock vina software. -

Download Author Version (PDF)
PCCP Accepted Manuscript This is an Accepted Manuscript, which has been through the Royal Society of Chemistry peer review process and has been accepted for publication. Accepted Manuscripts are published online shortly after acceptance, before technical editing, formatting and proof reading. Using this free service, authors can make their results available to the community, in citable form, before we publish the edited article. We will replace this Accepted Manuscript with the edited and formatted Advance Article as soon as it is available. You can find more information about Accepted Manuscripts in the Information for Authors. Please note that technical editing may introduce minor changes to the text and/or graphics, which may alter content. The journal’s standard Terms & Conditions and the Ethical guidelines still apply. In no event shall the Royal Society of Chemistry be held responsible for any errors or omissions in this Accepted Manuscript or any consequences arising from the use of any information it contains. www.rsc.org/pccp Page 1 of 11 PhysicalPlease Chemistry do not adjust Chemical margins Physics PCCP PAPER Effect of nanosize on surface properties of NiO nanoparticles for adsorption of Quinolin-65 ab a a Received 00th January 20xx, Nedal N. Marei, Nashaat N. Nassar* and Gerardo Vitale Accepted 00th January 20xx Using Quinolin-65 (Q-65) as a model-adsorbing compound for polar heavy hydrocarbons, the nanosize effect of NiO Manuscript DOI: 10.1039/x0xx00000x nanoparticles on adsorption of Q-65 was investigated. Different-sized NiO nanoparticles with sizes between 5 and 80 nm were prepared by controlled thermal dehydroxylation of Ni(OH)2. -

BIOVIA Discovery Studio
3DS.COM/BIOVIA3DS.COM/BIOVIA © © DassaultDassault Systèmes Systèmes| |Confidential Confidential InformationInformation | |3/16/2019 3/16/2019| BIOVIA Discovery Discovery BIOVIA COMPREHENSIVE MODELING 創源生技 FOR FOR SCIENCESLIFE ANDSIMULATIONS 經理 陳冠文 分子視算中心 Studio (Gene) 3DS.COM/BIOVIA © Dassault Systèmes | Confidential Information | 3/16/2019 | Copyright©2019 GGA Corp., All rights reserved. AllCorp., GGA Copyright and Disclaimer • Copyright © 2019 GGA corp. All rights reserved. • This presentation and/or any related documents contains statements regarding our plans or expectations | for future features, enhancements or functionalities of current or future products (collectively "Enhancements"). Our plans or expectations are subject to change at any time at our discretion. 3/16/2019 Accordingly, GGA Corp. is making no representation, undertaking no commitment or legal obligation to create, develop or license any product or Enhancements. • The presentation, documents or any related statements are not intended to, nor shall, create any legal | Confidential Information | Information | Confidential obligation upon GGA Corp., and shall not be relied upon in purchasing any product. Any such obligation shall only result from a written agreement executed by both parties. Systèmes • In addition, information disclosed in this presentation and related documents, whether oral or written, is © Dassault Dassault © confidential or proprietary information of GGA Corp.. It shall be used only for the purpose of furthering our business relationship, and shall not be disclosed to third parties. 3DS.COM/BIOVIA Copyright©2019 GGA Corp., All rights reserved. GGA is part of the BIONET Group (訊聯生物科技) | CEO: Christopher Tsai, Ph.D. 蔡政憲 博士 3/16/2019 Established: Nov. 2008 Main Product & Service Areas: | Confidential Information | Information | Confidential 1. -

Supporting Information
Electronic Supplementary Material (ESI) for Chemical Science. This journal is © The Royal Society of Chemistry 2015 Supporting Information A Single Crystalline Porphyrinic Titanium Metal−Organic Framework Shuai Yuana†, Tian-Fu Liua†, Dawei Fenga, Jian Tiana, Kecheng Wanga, Junsheng Qina, a a a a b Qiang Zhang , Ying-Pin Chen , Mathieu Bosch , Lanfang Zou , Simon J. Teat, Scott J. c a Dalgarno and Hong-Cai Zhou * a Department of Chemistry, Texas A&M University, College Station, Texas 77842-3012, USA b Advanced Light Source, Lawrence Berkeley National Laboratory Berkeley, CA 94720, USA c Institute of Chemical Sciences, Heriot-Watt University Riccarton, Edinburgh EH14 4AS, U.K. † Equal contribution to this work *To whom correspondence should be addressed. Email: [email protected] Tel: +1 (979) 845-4034; Fax: +1 (979) 845-1595 S1 Contents S1. Ligand Synthesis..............................................................................................................3 S2. Syntheses of PCN-22.......................................................................................................5 S3. X-ray Crystallography .....................................................................................................6 S4. Topological Analyses ......................................................................................................9 S5. N2 Sorption Isotherm .....................................................................................................10 S6. Simulation of the Accessible Surface Area ...................................................................12 -

Quantitative Structure-Activity Relationship and Molecular Docking
Journal of Advanced Research (2017) 8, 33–43 Cairo University Journal of Advanced Research ORIGINAL ARTICLE Quantitative structure-activity relationship and molecular docking studies of a series of quinazolinonyl analogues as inhibitors of gamma amino butyric acid aminotransferase Usman Abdulfatai *, Adamu Uzairu, Sani Uba Department of Chemistry, Ahmadu Bello University, P.M.B. 1044, Zaria, Nigeria GRAPHICAL ABSTRACT ARTICLE INFO ABSTRACT Article history: Quantitative structure-activity relationship and molecular docking studies were carried out on a Received 4 July 2016 series of quinazolinonyl analogues as anticonvulsant inhibitors. Density Functional Theory Received in revised form 11 October (DFT) quantum chemical calculation method was used to find the optimized geometry of the 2016 anticonvulsants inhibitors. Four types of molecular descriptors were used to derive a quantita- tive relation between anticonvulsant activity and structural properties. The relevant molecular * Corresponding author. Fax: +234 (+603) 6196 4053. E-mail address: [email protected] (U. Abdulfatai). Peer review under responsibility of Cairo University. Production and hosting by Elsevier http://dx.doi.org/10.1016/j.jare.2016.10.004 2090-1232 Ó 2016 Production and hosting by Elsevier B.V. on behalf of Cairo University. This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/). 34 U. Abdulfatai et al. Accepted 15 October 2016 descriptors were selected by Genetic Function Algorithm (GFA). The best model was validated Available online 16 November 2016 and found to be statistically significant with squared correlation coefficient (R2) of 0.934, 2 adjusted squared correlation coefficient (Radj) value of 0.912, Leave one out (LOO) cross valida- 2 2 Keywords: tion coefficient (Q ) value of 0.8695 and the external validation (Rpred) of 0.72. -

Preparing a PDB File the Protein Data Bank (PDB) Is Possibly the World’S Leading Public Source of Three-Dimensional Data for Biological Molecules (1)
Copyright ©2006, Accelrys Software Inc. All rights reserved. Preparing a PDB File The Protein Data Bank (PDB) is possibly the world’s leading public source of three-dimensional data for biological molecules (1). As of July 2006, over 37,000 entries could be found in the PDB. Hundreds more are being added every month. Both X-ray diffraction and other solid-state techniques account for the majority of the structures. However, over 5500 NMR structures are also available. These deposited structures include proteins, peptides, nucleic acids, carbohydrates, and complexes of these molecules. Figure 1: Schematic view of the ligand-binding domain from the As a first step in a modeling project, many vitamin D receptor (PDB file 1IE9). researchers look in the PDB to find available The crystallographic waters are shown structures related to their project. Preparation of as small spheres and the bound ligand these molecules for work in the Discovery Studio is shown as a CPK model. environment is a critical process to your modeling OH efforts. H3C CH In the following steps, we will load a PDB file for 3 the ligand-binding domain of the vitamin D receptor H C 3 (VDR) in complex with a ligand (named VDX in CH 3 this exercise). The file is 1IE9 as reported by Tocchini-Valentini et al. (5). The vitamin D receptor is responsible for the expression of a H variety of genes including calcium metabolism, bone formation, and cell growth and differentiation CH 2 (2). Understanding VDR conformational changes resulting from interactions with bound ligands may HO OH help to identify and treat persons at risk for Figure 2: 1α,25-dihydroxyvitamin disorders such as osteoporosis, breast cancer, or D3, the metabolized form of prostate cancer.