Cultivating the Uncultured: Growing the Recalcitrant Cluster-2 Frankia Strains
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Cally Plant List a ACIPHYLLA Horrida
Cally Plant List A ACIPHYLLA horrida ACONITUM albo-violaceum albiflorum ABELIOPHYLLUM distichum ACONITUM cultivar ABUTILON vitifolium ‘Album’ ACONITUM pubiceps ‘Blue Form’ ACAENA magellanica ACONITUM pubiceps ‘White Form’ ACAENA species ACONITUM ‘Spark’s Variety’ ACAENA microphylla ‘Kupferteppich’ ACONITUM cammarum ‘Bicolor’ ACANTHUS mollis Latifolius ACONITUM cammarum ‘Franz Marc’ ACANTHUS spinosus Spinosissimus ACONITUM lycoctonum vulparia ACANTHUS ‘Summer Beauty’ ACONITUM variegatum ACANTHUS dioscoridis perringii ACONITUM alboviolaceum ACANTHUS dioscoridis ACONITUM lycoctonum neapolitanum ACANTHUS spinosus ACONITUM paniculatum ACANTHUS hungaricus ACONITUM species ex. China (Ron 291) ACANTHUS mollis ‘Long Spike’ ACONITUM japonicum ACANTHUS mollis free-flowering ACONITUM species Ex. Japan ACANTHUS mollis ‘Turkish Form’ ACONITUM episcopale ACANTHUS mollis ‘Hollard’s Gold’ ACONITUM ex. Russia ACANTHUS syriacus ACONITUM carmichaelii ‘Spätlese’ ACER japonicum ‘Aconitifolium’ ACONITUM yezoense ACER palmatum ‘Filigree’ ACONITUM carmichaelii ‘Barker’s Variety’ ACHILLEA grandifolia ACONITUM ‘Newry Blue’ ACHILLEA ptarmica ‘Perry’s White’ ACONITUM napellus ‘Bergfürst’ ACHILLEA clypeolata ACONITUM unciniatum ACIPHYLLA monroi ACONITUM napellus ‘Blue Valley’ ACIPHYLLA squarrosa ACONITUM lycoctonum ‘Russian Yellow’ ACIPHYLLA subflabellata ACONITUM japonicum subcuneatum ACONITUM meta-japonicum ADENOPHORA aurita ACONITUM napellus ‘Carneum’ ADIANTUM aleuticum ‘Japonicum’ ACONITUM arcuatum B&SWJ 774 ADIANTUM aleuticum ‘Miss Sharples’ ACORUS calamus ‘Argenteostriatus’ -

The Wood Cross Sections of Hermann Nördlinger (1818–1897)
IAWA Journal, Vol. 29 (4), 2008: 439–457 THE WOOD CROSS SECTIONS OF HERMANN NÖRDLINGER (1818–1897) Ben Bubner Leibniz-Zentrum für Agrarlandschaftsforschung (ZALF) e.V., Institut für Landschaftsstoffdynamik, Eberswalder Str. 84, 15374 Müncheberg, Germany [E-mail: [email protected]] SUMMARY Hermann Nördlinger (1818–1897), forestry professor in Hohenheim, Germany, published a series of wood cross sections in the years 1852 to 1888 that are introduced here to the modern wood anatomist. The sec- tions, which vary from 50 to 100 μm in thickness, are mounted on sheets of paper and their quality is high enough to observe microscopic details. Their technical perfection is as remarkable as the mode of distribution: sections of 100 wood species were presented in a box together with a booklet containing wood anatomical descriptions. These boxes were dis- tributed as books by the publisher Cotta, from Stuttgart, Germany, with a maximum circulation of 500 per volume. Eleven volumes comprise 1100 wood species from all over the world. These include not only conifers and broadleaved trees but also shrubs, ferns and palms representing a wide variety of woody structures. Excerpts of this collection were also pub- lished in Russian, English and French. Today, volumes of Nördlingerʼs cross sections are found in libraries throughout Europe and the United States. Thus, they are relatively easily accessible to wood anatomists who are interested in historic wood sections. A checklist with the content of each volume is appended. Key words: Cross section, wood collection, wood anatomy, history. INTRODUCTION Wood scientists who want to distinguish wood species anatomically rely on thin sec- tions mounted on glass slides and descriptions in books that are illustrated with micro- photographs. -

Dr. Duke's Phytochemical and Ethnobotanical Databases List of Plants for Lyme Disease (Chronic)
Dr. Duke's Phytochemical and Ethnobotanical Databases List of Plants for Lyme Disease (Chronic) Plant Chemical Count Activity Count Garcinia xanthochymus 1 1 Nicotiana rustica 1 1 Acacia modesta 1 1 Galanthus nivalis 1 1 Dryopteris marginalis 2 1 Premna integrifolia 1 1 Senecio alpinus 1 1 Cephalotaxus harringtonii 1 1 Comptonia peregrina 1 1 Diospyros rotundifolia 1 1 Alnus crispa 1 1 Haplophyton cimicidum 1 1 Diospyros undulata 1 1 Roylea elegans 1 1 Bruguiera gymnorrhiza 1 1 Gmelina arborea 1 1 Orthosphenia mexicana 1 1 Lumnitzera racemosa 1 1 Melilotus alba 2 1 Duboisia leichhardtii 1 1 Erythroxylum zambesiacum 1 1 Salvia beckeri 1 1 Cephalotaxus spp 1 1 Taxus cuspidata 3 1 Suaeda maritima 1 1 Rhizophora mucronata 1 1 Streblus asper 1 1 Plant Chemical Count Activity Count Dianthus sp. 1 1 Glechoma hirsuta 1 1 Phyllanthus flexuosus 1 1 Euphorbia broteri 1 1 Hyssopus ferganensis 1 1 Lemaireocereus thurberi 1 1 Holacantha emoryi 1 1 Casearia arborea 1 1 Fagonia cretica 1 1 Cephalotaxus wilsoniana 1 1 Hydnocarpus anthelminticus 2 1 Taxus sp 2 1 Zataria multiflora 1 1 Acinos thymoides 1 1 Ambrosia artemisiifolia 1 1 Rhododendron schotense 1 1 Sweetia panamensis 1 1 Thymelaea hirsuta 1 1 Argyreia nervosa 1 1 Carapa guianensis 1 1 Parthenium hysterophorus 1 1 Rhododendron anthopogon 1 1 Strobilanthes cusia 1 1 Dianthus superbus 1 1 Pyropolyporus fomentarius 1 1 Euphorbia hermentiana 1 1 Porteresia coarctata 1 1 2 Plant Chemical Count Activity Count Aerva lanata 1 1 Rivea corymbosa 1 1 Solanum mammosum 1 1 Juniperus horizontalis 1 1 Maytenus -

They Come in Teams
GBE Frankia-Enriched Metagenomes from the Earliest Diverging Symbiotic Frankia Cluster: They Come in Teams Thanh Van Nguyen1, Daniel Wibberg2, Theoden Vigil-Stenman1,FedeBerckx1, Kai Battenberg3, Kirill N. Demchenko4,5, Jochen Blom6, Maria P. Fernandez7, Takashi Yamanaka8, Alison M. Berry3, Jo¨ rn Kalinowski2, Andreas Brachmann9, and Katharina Pawlowski 1,* 1Department of Ecology, Environment and Plant Sciences, Stockholm University, Sweden 2Center for Biotechnology (CeBiTec), Bielefeld University, Germany 3Department of Plant Sciences, University of California, Davis 4Laboratory of Cellular and Molecular Mechanisms of Plant Development, Komarov Botanical Institute, Russian Academy of Sciences, Saint Petersburg, Russia 5Laboratory of Molecular and Cellular Biology, All-Russia Research Institute for Agricultural Microbiology, Saint Petersburg, Russia 6Bioinformatics and Systems Biology, Justus Liebig University, Gießen, Germany 7Ecologie Microbienne, Centre National de la Recherche Scientifique UMR 5557, Universite Lyon I, Villeurbanne Cedex, France 8Forest and Forestry Products Research Institute, Ibaraki, Japan 9Biocenter, Ludwig Maximilians University Munich, Planegg-Martinsried, Germany *Corresponding author: E-mail: [email protected]. Accepted: July 10, 2019 Data deposition: This project has been deposited at EMBL/GenBank/DDBJ under the accession PRJEB19438 - PRJEB19449. Abstract Frankia strains induce the formation of nitrogen-fixing nodules on roots of actinorhizal plants. Phylogenetically, Frankia strains can be grouped in four clusters. The earliest divergent cluster, cluster-2, has a particularly wide host range. The analysis of cluster-2 strains has been hampered by the fact that with two exceptions, they could never be cultured. In this study, 12 Frankia-enriched meta- genomes of Frankia cluster-2 strains or strain assemblages were sequenced based on seven inoculum sources. Sequences obtained via DNA isolated from whole nodules were compared with those of DNA isolated from fractionated preparations enhanced in the Frankia symbiotic structures. -

A Global View on the Riparian Forests with Salix
This article was downloaded by: [Tech Univ of Lisbon Polo Ist] On: 11 July 2011, At: 08:56 Publisher: Taylor & Francis Informa Ltd Registered in England and Wales Registered Number: 1072954 Registered office: Mortimer House, 37-41 Mortimer Street, London W1T 3JH, UK Plant Biosystems - An International Journal Dealing with all Aspects of Plant Biology Publication details, including instructions for authors and subscription information: http://www.tandfonline.com/loi/tplb20 A global view on the riparian forests with Salix neotricha and Populus alba in the Iberian Peninsula (Portugal and Spain) José Carlos Costa a , Carlos Neto b , Jorge Capelo c , Mário Lousã a & Salvador Rivas-Martínez d a Departamento de Protecção de Plantas e de Fitoecologia, Instituto Superior de Agronomia, Technical University of Lisbon (TULisbon), Tapada da Ajuda, 1349-017, Lisboa, Portugal b Instituto de Geografia e Ordenamento do Território, Universidade de Lisboa, Alameda da Universidade, 1600-214, Lisboa, Portugal c Instituto Nacional de Recursos Biológicos, I. P., Quinta do Marquês, 2780-159, Oeiras, Portugal d Phytosociological Research Center (CIF), J. M. Usandizaga 46., E-28409 Los Negrales, Madrid, España, Spain Available online: 8 July 2011 To cite this article: José Carlos Costa, Carlos Neto, Jorge Capelo, Mário Lousã & Salvador Rivas-Martínez (2011): A global view on the riparian forests with Salix neotricha and Populus alba in the Iberian Peninsula (Portugal and Spain), Plant Biosystems - An International Journal Dealing with all Aspects of Plant Biology, DOI:10.1080/11263504.2011.584719 To link to this article: http://dx.doi.org/10.1080/11263504.2011.584719 PLEASE SCROLL DOWN FOR ARTICLE Full terms and conditions of use: http://www.tandfonline.com/page/terms-and-conditions This article may be used for research, teaching and private study purposes. -

Anti-Bacterial Activity of Coriaria Myrtifolia Against Agrobacterium Tumefaciens: Plant Pathogen Responsible for Crown Gall
Vol. 7(48), pp. 5529-5532, 4 December, 2013 DOI: 10.5897/AJMR2013.6284 ISSN 1996-0808 ©2013 Academic Journals African Journal of Microbiology Research http://www.academicjournals.org/AJMR Short Communication Anti-bacterial activity of Coriaria myrtifolia against Agrobacterium tumefaciens: Plant pathogen responsible for crown gall Halima BERRADA1, Abdellah FARAH2, Mouhcine FADIL3 and Kawtar FIKRI BENBRAHIM1* 1Laboratory of Microbial Biotechnology, Faculty of Sciences and Technology, Sidi Mohammed Ben Abdellah University, P. O. Box 2202, Fez, Morocco. 2Laboratory of Medicinal, Aromatic Plants and Natural Substances in the National Institute of Medicinal and Aromatic Plants, Taounate, Morocco. 3Laboratory of Functional Ecology and Environment, Faculty of Sciences and Technology, Sidi Mohammed Ben Abdellah University, P. O. Box 2202, Fez, Morocco. Accepted 11 November, 2013 The present work aimed to evaluate the antibacterial activity of aqueous and methanolic extracts of Coriaria myrtifolia’s leaves against Agrobacterium sp. and Agrobacterium tumefaciens “plant pathogen responsible for crown gall” in an objective to identify novel antimicrobial agents and to put forward efforts of proving plant’s extracts scientific credibility, and determining their spectrum of activity. The bacteria tested were found profoundly sensitive to both of the C. myrtifolia extracts. The extent of inhibition was more important by methanolic extract than by aqueous one. The average diameter of inhibition zones ranged from 10.67 to 15.33 mm and 12.68 to 18 mm for aqueous and methanolic extract, respectively. This study was the first to report the antimicrobial activity of extracts obtained from the leaf of C. myrtifolia against Agrobacterium sp. and Agrobacterium tumefaciens. It can be concluded that the observed antibacterial characteristics of C. -

Economically Important Plants Arranged Systematically James P
Humboldt State University Digital Commons @ Humboldt State University Botanical Studies Open Educational Resources and Data 1-2017 Economically Important Plants Arranged Systematically James P. Smith Jr Humboldt State University, [email protected] Follow this and additional works at: http://digitalcommons.humboldt.edu/botany_jps Part of the Botany Commons Recommended Citation Smith, James P. Jr, "Economically Important Plants Arranged Systematically" (2017). Botanical Studies. 48. http://digitalcommons.humboldt.edu/botany_jps/48 This Economic Botany - Ethnobotany is brought to you for free and open access by the Open Educational Resources and Data at Digital Commons @ Humboldt State University. It has been accepted for inclusion in Botanical Studies by an authorized administrator of Digital Commons @ Humboldt State University. For more information, please contact [email protected]. ECONOMICALLY IMPORTANT PLANTS ARRANGED SYSTEMATICALLY Compiled by James P. Smith, Jr. Professor Emeritus of Botany Department of Biological Sciences Humboldt State University Arcata, California 30 January 2017 This list began in 1970 as a handout in the Plants and Civilization course that I taught at HSU. It was an updating and expansion of one prepared by Albert F. Hill in his 1952 textbook Economic Botany... and it simply got out of hand. I also thought it would be useful to add a brief description of how the plant is used and what part yields the product. There are a number of more or less encyclopedic references on this subject. The number of plants and the details of their uses is simply overwhelming. In the list below, I have attempted to focus on those plants that are of direct economic importance to us. -

Ethnobotanical Studies in Western Gironès (Catalonia, Iberian Peninsula) Airy Gras1,2,3, Ginesta Serrasolses1, Joan Vallès1,3,4 and Teresa Garnatje2*
Gras et al. Journal of Ethnobiology and Ethnomedicine (2019) 15:19 https://doi.org/10.1186/s13002-019-0295-2 RESEARCH Open Access Traditional knowledge in semi-rural close to industrial areas: ethnobotanical studies in western Gironès (Catalonia, Iberian Peninsula) Airy Gras1,2,3, Ginesta Serrasolses1, Joan Vallès1,3,4 and Teresa Garnatje2* Abstract Background: The western Gironès is a district located in NE Catalonia (NE Iberian Peninsula). This area comprising 186.55 km2 and 10,659 inhabitants is composed of 5 municipalities encompassing 29 villages, located in the hydrographic basins of the Ter and Llémena rivers. Methods: Following the methodology based on the semi-structured interviews, we carried out 40 interviews with 57 informants, 31 were women and the remaining 26 were men, with an average age of 78.6 years. Results: In the present study, data from 316 taxa (301 angiosperms, 8 gymnosperms, and 7 pteridophytes) belonging to 89 botanical families were collected. The interviewed informants referred 3776 UR of 298 taxa, 1933 (51.19%) of them corresponding to the food category, 949 (25.13%) to the medicinal ones, and 894 (23.68%) to other uses. In addition, 581 vernacular names for 306 species, subspecies, and varieties have also been collected. Conclusions: These results reveal the validity of traditional knowledge in the studied area, which can be seriously threatened by the loss of its rural condition and its proximity to industrialized areas. Keywords: Ethnobotany, Ethnoflora, Gironès, Medicinal uses, Plant uses, Traditional knowledge Introduction For this reason, the research that was initially focused The Catalan-speaking territories constitute a cultural on non-industrialized areas [16–22] has now been ex- unity that has attracted the interest of researchers from panded in industrialized areas due to their rapid loss of various disciplines. -

Early Evolution of Coriariaceae (Cucurbitales) in Light of a New Early Campanian (Ca
TAXON 69 (1) • February 2020: 87–99 Renner & al. • Evolution of Coriariaceae SYSTEMATICS AND PHYLOGENY Early evolution of Coriariaceae (Cucurbitales) in light of a new early Campanian (ca. 82 Mya) pollen record from Antarctica Susanne S. Renner,1 Viviana D. Barreda,2 María Cristina Tellería,3 Luis Palazzesi2 & Tanja M. Schuster1 1 Systematic Botany and Mycology, Ludwig Maximilian University of Munich (LMU), Menzinger-Straße 67, 80638 Munich, Germany 2 Museo Argentino de Ciencias Naturales, Av. Ángel Gallardo 470, C1407DJR Buenos Aires, Argentina 3 Laboratorio de Sistemática y Biología Evolutiva, Museo de La Plata, La Plata, B1900FWA, Argentina Address for correspondence: Susanne S. Renner, [email protected] DOI https://doi.org/10.1002/tax.12203 Abstract Coriariaceae comprise only Coriaria, a genus of shrubs with nine species in Australasia (but excluding Australia), five in the Himalayas, Taiwan, the Philippines, and Japan, one in the Mediterranean, and one ranging from Patagonia to Mexico. The sister family, Corynocarpaceae, comprises five species of evergreen trees from New Guinea to New Zealand and Australia. This distribution has long fascinated biogeographers as potential support for Wegener’s theory of continental drift, with alternative scenarios invoking either Antarctic or Beringian range expansions. Here, we present the discovery of pollen grains from Early Campanian (ca. 82 Mya) deposits in Antarctica, which we describe as Coriaripites goodii sp. nov., and newly generated nuclear and plastid molecular data for most of the family’s species and its outgroup. This greatly expands the family’s fossil record and is the so far oldest fossil of the order Cucurbitales. We used the phylogeny, new fossil, and an Oligocene flowering branch assigned to a small subclade of Coriaria to gen- erate a chronogram and to study changes in chromosome number, deciduousness, and andromonoecy. -
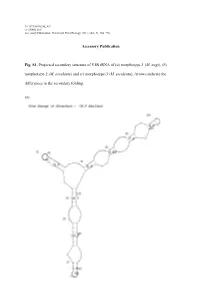
Accessory Publication Fig. S1. Projected Secondary Structure of 5.8
10.1071/FP10248_AC © CSIRO 2011 Accessory Publication: Functional Plant Biology, 2011, 38(8–9), 768–776. Accessory Publication Fig. S1. Projected secondary structure of 5.8S rRNA of (a) morphotype-1 (M. nagi), (b) morphotype-2 (M. esculenta) and (c) morphotype-3 (M. esculenta). Arrows indicate the differences in the secondary folding. (a) (b) (c) Table S1. GenBank accession numbers of nucleotide sequences used. GENBANK Sl. NAME OF THE GENUS AND REGION ACCESSION REFERENCE No. NUMBER 1. Myrica nagi (Morphotype-1, ME1) ITS FJ469992 Present study 2. M. esculenta (Morphotype-2, ME2) ITS FJ469993 Present study 3. M. esculenta (Morphotype-3, ME3) ITS FJ469994 Present study 4. M. nagi (Mophotype-1, MYR1) 18S rDNA FJ469989 Present study 5. M. esculenta (Morphotype-2, MYR2) 18S rDNA FJ569990 Present study 6. M. esculenta (Morphotype-3, MYR3) 18S rDNA FJ469991 Present study 7. Alnus cordata ITS AY352306 Chen and Li (2004) 8. A. glutinosa ITS AY352310 Chen and Li (2004) 9. A. japonica ITS AY352314 Chen and Li (2004) 10. A. orientalis ITS AY352320 Chen and Li (2004) 11. A. serrulata ITS AY352322 Chen and Li (2004) 12. A. sibirica ITS AY352323 Chen and Li (2004) 13. A. tenuifolia ITS AY352327 Chen and Li (2004) 14. A. viridis ITS AY352329 Chen and Li (2004) 15. Berberidopsis corallina 18S rDNA AF206866 Soltis et al. (1999) (unpublished) 16. Betula humilis ITS AJ783643 Forest et al. (2005) 17. B. nana ITS AY352336 Chen and Li (2004) 18. B. uber ITS AY352334 Chen and Li (2004) 19. Carya glabra 18S rDNA AF206880 Soltis et al. (1999) (unpublished) 20. -

Antioxidant and Anti-Inflammatory Activities Evaluation of Coriaria Myrtifolia from the North of Morocco
International Food Research Journal 24(2): 498-502 (April 2017) Journal homepage: http://www.ifrj.upm.edu.my Antioxidant and anti-inflammatory activities evaluation of Coriaria myrtifolia from the North of Morocco 1,2Hafsé, M., 2,3Farah, A., 4Mouktadir, J. E. and 1*Fikri-Benbrahim, K. 1Microbial Biotechnology Laboratory. Faculty of Science and Technology. Saiss. Sidi Mohamed Ben Abdellah University. Fez. Morocco 2National Institute of Medicinal and Aromatic Plants. Taounate. Morocco 3Applied Organic Chemistry Laboratory, Faculty of Sciences and Techniques, Sidi Mohamed Ben Abdellah University, P. O. Box 2202 Imouzzer Road, Fez, Morocco. 4Discourse, Society and Creativity: Perception and Implications Laboratory. Faculty of Arts and Humanities Saiss. Sidi Mohamed Ben Abdellah University. Fez. Morocco Article history Abstract Received: 1 January 2016 The present study has examined the chemical composition and evaluated the pharmacological Received in revised form: activities of the ethyl acetate extract of Coriaria myrtifolia. The antioxidant activity was 17 April 2016 Accepted: 21 April 2016 determined by using the diphenyl-picryl-hydrazyl (DPPH) test, and the anti-inflammatory activity was evaluated by using the plantar edema model induced in rabbits by carrageenan. The extract has revealed a significant free-radical scavenging capacity. The plant IC50 value of Keywords 0.016 mg/ml was found less than stated with the Butylated hydroxyl-toluene (BHT) standard (0.025 mg/ml). Administration of the ethyl acetate extract at a dose of 0.013mg/kg wb inhibits Coriaria myrtifolia Chemical composition completely the inflammation. These results have indicated that the C. myrtifolia leaves Antioxidant activity contains bioactive compounds and have a high antioxidant activity as well as interesting anti- Anti-inflammatory activity inflammatory properties, suggesting the usefulness of their extracts to prevent oxidative and inflammatory processes. -

Anthocyanin Composition of the Fruit of Coriaria Myrtifolia L
PHYTOCHEMICAL ANALYSIS Phytochem. Anal. 13, 354–357 (2002) Published online in Wiley InterScience (www.interscience.wiley.com). DOI: 10.1002/pca.667 Anthocyanin Composition of the Fruit of Coriaria myrtifolia L. Marı´a Teresa Escribano-Bailo´n,1 Celestino Santos-Buelga,1 Gonzalo L. Alonso2 and Marı´a Rosario Salinas2* 1Laboratorio de Nutricio´n y Bromatologı´a, Facultad de Farmacia, Universidad de Salamanca, E-37007 Salamanca, Spain 2Ca´tedra de Quı´mica Agrı´cola, ETSI Agro´nomos, Universidad de Castilla-La Mancha, E-02071 Albacete, Spain The anthocyanin composition of the fruit of Coriaria myrtifolia L. and the changes which occur during ripening were studied using HPLC-PAD and LC-MS. Ten anthocyanins were detected and identified by their absorption and mass spectra as the 3-glucoside and 3-galactoside derivatives of delphinidin, cyanidin, petunidin, peonidin and malvidin. Fruit ripening was accompanied by substantial changes in the anthocyanin profile, with methoxylated anthocyanins, i.e. malvidin and peonidin, predominating in the final stages of ripening, and the trihydroxylated anthocyanin, delphinidin, during the earlier stages. Furthermore, galactoside derivatives were more abundant than glucosides in the ripe fruit. At full maturity, the fruits of C. myrtifolia were very rich in anthocyanins with a content of 10.7% (on a dry weight basis), a level which is higher than that found in most fruits usually considered to be anthocyanin-rich. The ability to grow C. myrtifolia in damaged and nitrogen poor soils, together with the possibility of using this plant for the extraction of anthocyanin, makes it ideal for consolidating soils and repopulating semi-desert or fire-damaged areas.