(Poaceae) by Ekaterina (Katya) Boudko, B.Sc
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Taxonomical Notes on the Genus Piptatherum P. Beauv
TAXONOMICAL NOTES ON THE GENUS PIPTATHERUM P. BEAUV. (POACEAE) IN IRAN ee & M. Assadi׳B. Hamzeh Received 2015. 02. 20; accepted for publication 2015. 05. 12 ee, B. & Assadi, M. 2015. 06. 30: Taxonomical notes on the genus Piptatherum P. Beauv. (Poaceae) in׳Hamzeh Iran. –Iran. J. Bot. 21 (1): 01-09. Tehran. Piptatherum denaense is described as a new species from south west of Iran, Dena Mountain. It is close to P. laterale but differs from it by having glabrous lemma in lower half and toward the apex, narrower vegetative shoots and unbearded anthers. Piptatherum holciforme subsp. holciforme var. glabrum is not accepted as a distinct variety. Piptatherum sphacelatum formerly known as a synonym of P. molinioides is established as a separate species due to having distinct morphological characters as well as molecular differences based on literature. Thus, the number of taxa in the genus Piptatherum changes to 9 species, 2 subspecies and 4 varieties in Iran. An identification key to Piptatherum taxa occuring in Iran is provided. The distribution map of P. denaense with P. laterale subsp. laterale and the illustration of the new species are included. ee (correspondence <[email protected]>) & Mostafa Assadi, Research Institute of Forests and׳Behnam Hamzeh Rangelands, P. O. Box: 13185-116, Tehran, Iran. Key words: Piptatherum; Poaceae; new species; new synonym; reestablished species; Iran ﻧﻜﺎﺗﻲ در ﻣﻮرد ﺟﻨﺲ .Poaceae ) Piptatherum P. Beauv) در اﻳﺮان ﺑﻬﻨﺎم ﺣﻤﺰة، اﺳﺘﺎدﻳﺎر ﭘﮋوﻫﺶ ﻣﺆﺳﺴﻪ ﺗﺤﻘﻴﻘﺎت ﺟﻨﮕﻠﻬﺎ و ﻣﺮاﺗﻊ ﻛﺸﻮر ﻣﺼﻄﻔﻲ اﺳﺪي، اﺳﺘﺎد ﭘﮋوﻫﺶ ﻣﺆﺳﺴﻪ ﺗﺤﻘﻴﻘﺎت ﺟﻨﮕﻠﻬﺎ و ﻣﺮاﺗﻊ ﻛﺸﻮر Piptatherum denaense ﺑﻪﻋﻨﻮان ﮔﻮﻧﻪاي ﺟﺪﻳﺪ از ﻛﻮه دﻧﺎ در ﺟﻨﻮب ﻏﺮب اﻳﺮان ﺷﺮح داده ﻣﻲﺷﻮد. -

Types of American Grasses
z LIBRARY OF Si AS-HITCHCOCK AND AGNES'CHASE 4: SMITHSONIAN INSTITUTION UNITED STATES NATIONAL MUSEUM oL TiiC. CONTRIBUTIONS FROM THE United States National Herbarium Volume XII, Part 3 TXE&3 OF AMERICAN GRASSES . / A STUDY OF THE AMERICAN SPECIES OF GRASSES DESCRIBED BY LINNAEUS, GRONOVIUS, SLOANE, SWARTZ, AND MICHAUX By A. S. HITCHCOCK z rit erV ^-C?^ 1 " WASHINGTON GOVERNMENT PRINTING OFFICE 1908 BULLETIN OF THE UNITED STATES NATIONAL MUSEUM Issued June 18, 1908 ii PREFACE The accompanying paper, by Prof. A. S. Hitchcock, Systematic Agrostologist of the United States Department of Agriculture, u entitled Types of American grasses: a study of the American species of grasses described by Linnaeus, Gronovius, Sloane, Swartz, and Michaux," is an important contribution to our knowledge of American grasses. It is regarded as of fundamental importance in the critical sys- tematic investigation of any group of plants that the identity of the species described by earlier authors be determined with certainty. Often this identification can be made only by examining the type specimen, the original description being inconclusive. Under the American code of botanical nomenclature, which has been followed by the author of this paper, "the nomenclatorial t}rpe of a species or subspecies is the specimen to which the describer originally applied the name in publication." The procedure indicated by the American code, namely, to appeal to the type specimen when the original description is insufficient to identify the species, has been much misunderstood by European botanists. It has been taken to mean, in the case of the Linnsean herbarium, for example, that a specimen in that herbarium bearing the same name as a species described by Linnaeus in his Species Plantarum must be taken as the type of that species regardless of all other considerations. -
Qrno. 1 2 3 4 5 6 7 1 CP 2903 77 100 0 Cfcl3
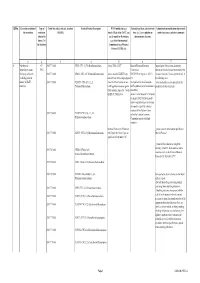
QRNo. General description of Type of Tariff line code(s) affected, based on Detailed Product Description WTO Justification (e.g. National legal basis and entry into Administration, modification of previously the restriction restriction HS(2012) Article XX(g) of the GATT, etc.) force (i.e. Law, regulation or notified measures, and other comments (Symbol in and Grounds for Restriction, administrative decision) Annex 2 of e.g., Other International the Decision) Commitments (e.g. Montreal Protocol, CITES, etc) 12 3 4 5 6 7 1 Prohibition to CP 2903 77 100 0 CFCl3 (CFC-11) Trichlorofluoromethane Article XX(h) GATT Board of Eurasian Economic Import/export of these ozone destroying import/export ozone CP-X Commission substances from/to the customs territory of the destroying substances 2903 77 200 0 CF2Cl2 (CFC-12) Dichlorodifluoromethane Article 46 of the EAEU Treaty DECISION on August 16, 2012 N Eurasian Economic Union is permitted only in (excluding goods in dated 29 may 2014 and paragraphs 134 the following cases: transit) (all EAEU 2903 77 300 0 C2F3Cl3 (CFC-113) 1,1,2- 4 and 37 of the Protocol on non- On legal acts in the field of non- _to be used solely as a raw material for the countries) Trichlorotrifluoroethane tariff regulation measures against tariff regulation (as last amended at 2 production of other chemicals; third countries Annex No. 7 to the June 2016) EAEU of 29 May 2014 Annex 1 to the Decision N 134 dated 16 August 2012 Unit list of goods subject to prohibitions or restrictions on import or export by countries- members of the -

State of New York City's Plants 2018
STATE OF NEW YORK CITY’S PLANTS 2018 Daniel Atha & Brian Boom © 2018 The New York Botanical Garden All rights reserved ISBN 978-0-89327-955-4 Center for Conservation Strategy The New York Botanical Garden 2900 Southern Boulevard Bronx, NY 10458 All photos NYBG staff Citation: Atha, D. and B. Boom. 2018. State of New York City’s Plants 2018. Center for Conservation Strategy. The New York Botanical Garden, Bronx, NY. 132 pp. STATE OF NEW YORK CITY’S PLANTS 2018 4 EXECUTIVE SUMMARY 6 INTRODUCTION 10 DOCUMENTING THE CITY’S PLANTS 10 The Flora of New York City 11 Rare Species 14 Focus on Specific Area 16 Botanical Spectacle: Summer Snow 18 CITIZEN SCIENCE 20 THREATS TO THE CITY’S PLANTS 24 NEW YORK STATE PROHIBITED AND REGULATED INVASIVE SPECIES FOUND IN NEW YORK CITY 26 LOOKING AHEAD 27 CONTRIBUTORS AND ACKNOWLEGMENTS 30 LITERATURE CITED 31 APPENDIX Checklist of the Spontaneous Vascular Plants of New York City 32 Ferns and Fern Allies 35 Gymnosperms 36 Nymphaeales and Magnoliids 37 Monocots 67 Dicots 3 EXECUTIVE SUMMARY This report, State of New York City’s Plants 2018, is the first rankings of rare, threatened, endangered, and extinct species of what is envisioned by the Center for Conservation Strategy known from New York City, and based on this compilation of The New York Botanical Garden as annual updates thirteen percent of the City’s flora is imperiled or extinct in New summarizing the status of the spontaneous plant species of the York City. five boroughs of New York City. This year’s report deals with the City’s vascular plants (ferns and fern allies, gymnosperms, We have begun the process of assessing conservation status and flowering plants), but in the future it is planned to phase in at the local level for all species. -

Taxonomy, Morphology and Palynology of Aegilops Vavilovii (Zhuk.) Chennav
African Journal of Agricultural Research Vol. 5(20), pp. 2841-2849, 18 October, 2010 Available online at http://www.academicjournals.org/AJAR ISSN 1991-637X ©2010 Academic Journals Full Length Research Paper Taxonomy, morphology and palynology of Aegilops vavilovii (Zhuk.) Chennav. (Poaceae: Triticeae) Evren Cabi1* Musa Doan1 Hülya Özler2, Galip Akaydin3 and Alptekin Karagöz4 1Department of Biological Sciences, Faculty of Arts and Sciences, Middle East Technical University, Ankara, Turkey. 2Department of Biology, Faculty of Arts and Sciences, Sinop University, Sinop Turkey. 3Department of Biology Education, Hacettepe University, 06800 Ankara, Turkey. 4Department of Biology, Aksaray University, Aksaray, Turkey. Accepted 23 September, 2010 Aegilops vavilovii (Zhuk.) Chennav., a rare species, was collected from Southeast Anatolia, Turkey. During the field studies of the project “Taxonomic revision of Tribe Triticeae in Turkey”, Ae. vavilovii was accidentally recollected from three localities in anliurfa and Mardin provinces in 2007 and 2008, respectively. The main objective of this study is to shed light on the diagnostic characteristics of this rare species including its morphological, palynological and micro morphological features. Moreover, an emended and expanded description, distribution, phenology and ecology of this rare species are also provided. A. vavilovii and A. crassa are naturally found in the Southeastern part of Turkey and they share similar morphological features that caused a confused taxonomy. Pollen grains of A. vavilovii are heteropolar, monoporate and spheroidal (A/B: 1,13) typically as Poaceous. However, it generally, prefers clayish loam soils that are slightly alkaline (pH 7.7) with low organic content (1.54%). Although it is a rare species with very narrow area of distribution, very few samples have been represented in ex situ collections and the species has not been involved in any in situ conservation activities to save its genetic resources in Turkey. -

Molecular Phylogenetic Analysis Resolves Trisetum
Journal of Systematics JSE and Evolution doi: 10.1111/jse.12523 Research Article Molecular phylogenetic analysis resolves Trisetum (Poaceae: Pooideae: Koeleriinae) polyphyletic: Evidence for a new genus, Sibirotrisetum and resurrection of Acrospelion Patricia Barberá1,3*,RobertJ.Soreng2 , Paul M. Peterson2* , Konstantin Romaschenko2 , Alejandro Quintanar1, and Carlos Aedo1 1Department of Biodiversity and Conservation, Real Jardín Botánico, CSIC, Madrid 28014, Spain 2Department of Botany, National Museum of Natural History, Smithsonian Institution, Washington DC 20013‐7012, USA 3Department of Africa and Madagascar, Missouri Botanical Garden, St. Louis, MO 63110, USA *Authors for correspondence. Patricia Barberá. E‐mail: [email protected]; Paul M. Peterson. E‐mail: [email protected] Received 4 March 2019; Accepted 5 May 2019; Article first published online 22 June 2019 Abstract To investigate the evolutionary relationships among the species of Trisetum and other members of subtribe Koeleriinae, a phylogeny based on DNA sequences from four gene regions (ITS, rpl32‐trnL spacer, rps16‐trnK spacer, and rps16 intron) is presented. The analyses, including type species of all genera in Koeleriinae (Acrospelion, Avellinia, Cinnagrostis, Gaudinia, Koeleria, Leptophyllochloa, Limnodea, Peyritschia, Rostraria, Sphenopholis, Trisetaria, Trisetopsis, Trisetum), along with three outgroups, confirm previous indications of extensive polyphyly of Trisetum. We focus on the monophyletic Trisetum sect. Sibirica cladethatweinterprethereasadistinctgenus,Sibirotrisetum gen. nov. We include adescriptionofSibirotrisetum with the following seven new combinations: Sibirotrisetum aeneum, S. bifidum, S. henryi, S. scitulum, S. sibiricum, S. sibiricum subsp. litorale,andS. turcicum; and a single new combination in Acrospelion: A. distichophyllum. Trisetum s.s. is limited to one, two or three species, pending further study. Key words: Acrospelion, Aveneae, grasses, molecular systematics, Poeae, Sibirotrisetum, taxonomy, Trisetum. -

Greene's Tuctoria 0 12.5 25 50 75 100
14. TUCTORIA GREENEI (GREENE’S TUCTORIA) a. Description and Taxonomy Taxonomy.—The genus Tuctoria is in the grass family (Poaceae), subfamily Chloridoideae, and is a member of the Orcuttieae tribe, which also includes Neostapfia and Orcuttia (Reeder 1965, Keeley 1998). Vasey (1891:146) originally assigned the name Orcuttia greenei to this species, from a type specimen collected in 1890 “on moist plains of the upper Sacramento, near Chico, California,” presumably in Butte County (Hoover 1941, Crampton 1958). Citing differences in lemma morphology, arrangement of the spikelets, and other differences (see “Description” below), Reeder (1982) segregated the genus Tuctoria from Orcuttia and created the new scientific name Tuctoria greenei for this species. Subsequent research suggests that Tuctoria is intermediate in evolutionary position between the primitive genus Neostapfia and the advanced genus Orcuttia (Keeley 1998, L. Boykin in litt. 2000). Several other common names have been used for this species, including Chico grass (Scribner 1899), awnless Orcutt grass (Abrams 1940), Greene’s orcuttia (Smith et al. 1980), and Greene’s Orcutt grass (California Department of Fish and Game 1991, U.S. Fish and Wildlife Service 1985c). Description and Identification.—The basic characteristics pertaining to all members of the Orcuttieae were described above in the Neostapfia colusana account. The genus Tuctoria is characterized by flattened spikelets similar to those of Orcuttia species, except that the spikelets of Tuctoria grow in a spiral, as opposed to a distichous, arrangement. Tuctoria species have short-toothed, narrow lemmas. The juvenile and terrestrial leaves of Tuctoria are similar to those of Orcuttia, but Tuctoria does not produce the floating type of intermediate leaves (Reeder 1982, Keeley 1998). -

Nature Conservation
J. Nat. Conserv. 11, – (2003) Journal for © Urban & Fischer Verlag http://www.urbanfischer.de/journals/jnc Nature Conservation Constructing Red Numbers for setting conservation priorities of endangered plant species: Israeli flora as a test case Yuval Sapir1*, Avi Shmida1 & Ori Fragman1,2 1 Rotem – Israel Plant Information Center, Dept. of Evolution, Systematics and Ecology,The Hebrew University, Jerusalem, 91904, Israel; e-mail: [email protected] 2 Present address: Botanical Garden,The Hebrew University, Givat Ram, Jerusalem 91904, Israel Abstract A common problem in conservation policy is to define the priority of a certain species to invest conservation efforts when resources are limited. We suggest a method of constructing red numbers for plant species, in order to set priorities in con- servation policy. The red number is an additive index, summarising values of four parameters: 1. Rarity – The number of sites (1 km2) where the species is present. A rare species is defined when present in 0.5% of the area or less. 2. Declining rate and habitat vulnerability – Evaluate the decreasing rate in the number of sites and/or the destruction probability of the habitat. 3. Attractivity – the flower size and the probability of cutting or exploitation of the plant. 4. Distribution type – scoring endemic species and peripheral populations. The plant species of Israel were scored for the parameters of the red number. Three hundred and seventy (370) species, 16.15% of the Israeli flora entered into the “Red List” received red numbers above 6. “Post Mortem” analysis for the 34 extinct species of Israel revealed an average red number of 8.7, significantly higher than the average of the current red list. -

Checklist of the Vascular Plants of Redwood National Park
Humboldt State University Digital Commons @ Humboldt State University Botanical Studies Open Educational Resources and Data 9-17-2018 Checklist of the Vascular Plants of Redwood National Park James P. Smith Jr Humboldt State University, [email protected] Follow this and additional works at: https://digitalcommons.humboldt.edu/botany_jps Part of the Botany Commons Recommended Citation Smith, James P. Jr, "Checklist of the Vascular Plants of Redwood National Park" (2018). Botanical Studies. 85. https://digitalcommons.humboldt.edu/botany_jps/85 This Flora of Northwest California-Checklists of Local Sites is brought to you for free and open access by the Open Educational Resources and Data at Digital Commons @ Humboldt State University. It has been accepted for inclusion in Botanical Studies by an authorized administrator of Digital Commons @ Humboldt State University. For more information, please contact [email protected]. A CHECKLIST OF THE VASCULAR PLANTS OF THE REDWOOD NATIONAL & STATE PARKS James P. Smith, Jr. Professor Emeritus of Botany Department of Biological Sciences Humboldt State Univerity Arcata, California 14 September 2018 The Redwood National and State Parks are located in Del Norte and Humboldt counties in coastal northwestern California. The national park was F E R N S established in 1968. In 1994, a cooperative agreement with the California Department of Parks and Recreation added Del Norte Coast, Prairie Creek, Athyriaceae – Lady Fern Family and Jedediah Smith Redwoods state parks to form a single administrative Athyrium filix-femina var. cyclosporum • northwestern lady fern unit. Together they comprise about 133,000 acres (540 km2), including 37 miles of coast line. Almost half of the remaining old growth redwood forests Blechnaceae – Deer Fern Family are protected in these four parks. -

Towards Preserving Threatened Grassland Species and Habitats
Towards preserving threatened grassland plant species and habitats - seed longevity, seed viability and phylogeography Dissertation zur Erlangung des Doktorgrades der Naturwissenschaften (Dr. rer. nat.) der Fakultät für Biologie und Vorklinische Medizin der Universität Regensburg vorgelegt von SIMONE B. TAUSCH aus Burghausen im Jahr 2017 II Das Promotionsgesuch wurde eingereicht am: 15.12.2017 Die Arbeit wurde angeleitet von: Prof. Dr. Peter Poschlod Regensburg, den 14.12.2017 Simone B. Tausch III IV Table of contents Chapter 1 General introduction 6 Chapter 2 Towards the origin of Central European grasslands: glacial and postgla- 12 cial history of the Salad Burnet (Sanguisorba minor Scop.) Chapter 3 A habitat-scale study of seed lifespan in artificial conditions 28 examining seed traits Chapter 4 Seed survival in the soil and at artificial storage: Implications for the 42 conservation of calcareous grassland species Chapter 5 How precise can X-ray predict the viability of wild flowering plant seeds? 56 Chapter 6 Seed dispersal in space and time - origin and conservation of calcareous 66 grasslands Summary 70 Zusammenfassung 72 References 74 Danksagung 89 DECLARATION OF MANUSCRIPTS Chapter 2 was published with the thesis’ author as main author: Tausch, S., Leipold, M., Poschlod, P. and Reisch, C. (2017). Molecular markers provide evidence for a broad-fronted recolonisation of the widespread calcareous grassland species Sanguisorba minor from southern and cryptic northern refugia. Plant Biology, 19: 562–570. doi:10.1111/plb.12570. V CHAPTER 1 General introduction THREATENED AND ENDANGERED persal ability (von Blanckenhagen & Poschlod, 2005). But in general, soils of calcareous grasslands exhibit HABITATS low ability to buffer species extinctions by serving as donor (Thompson et al., 1997; Bekker et al., 1998a; Regarding the situation of Europe’s plant species in- Kalamees & Zobel, 1998; Poschlod et al., 1998; Stöck- ventory, Central Europe represents the centre of en- lin & Fischer, 1999; Karlik & Poschlod, 2014). -

Final Report (Years 1-5) July 2007
Darwin Initiative: Project: 162 / 11 / 025 Cross-border conservation strategies for Altai Mountain endemics (Russia, Mongolia, Kazakhstan) Final Report (Years 1-5) July 2007 CONTENTS: DARWIN PROJECT INFORMATION 3 1 PROJECT BACKGROUND/RATIONALE 3 2 PROJECT SUMMARY 4 3 SCIENTIFIC, TRAINING, AND TECHNICAL ASSESSMENT 8 3.1 RESEARCH 8 Methodology 8 Liaison with local authorities and Regional Ecological Committees 9 Data storage and analysis 10 Results 11 3.2 TRAINING AND CAPACITY BUILDING ACTIVITIES. 14 4 PROJECT IMPACTS 15 5 PROJECT OUTPUTS 18 6 PROJECT EXPENDITURE 19 7 PROJECT OPERATION AND PARTNERSHIPS 19 8 ACTIONS TAKEN IN RESPONSE TO ANNUAL REPORT REVIEWS (IF APPLICABLE) 22 9 DARWIN IDENTITY 23 Project 162 / 11 / 025: Altai Mountains. Final Report, August 2007 1 10 LEVERAGE 23 11 SUSTAINABILITY AND LEGACY 24 12 VALUE FOR MONEY 25 APPENDIX I: PROJECT CONTRIBUTION TO ARTICLES UNDER THE CONVENTION ON BIOLOGICAL DIVERSITY (CBD) 27 APPENDIX II: OUTPUTS 29 APPENDIX III: PUBLICATIONS 35 APPENDIX IV: DARWIN CONTACTS 42 APPENDIX V: LOGICAL FRAMEWORK 44 APPENDIX VI: SELECTED TABLES 45 APPENDIX VII: COPIES OF INFORMATION LEAFLETS 52 APPENDIX VIII: PUBLICATIONS WITH SUMMARIES IN ENGLISH 53 APPENDIX IX: COPIES OF OUTPUTS SUPPLIED AS PDF 68 Project 162 / 11 / 025: Altai Mountains. Final Report, August 2007 2 Darwin Initiative for the Survival of Species Final Report Darwin Project Information Project Reference No. 162 / 11 / 025 Project Title Cross-border conservation strategies for Altai Mountain Endemics (Russia, Mongolia, Kazakhstan) Country(ies) UK, Russia, Mongolia, Kazakhstan UK Contractor University of Sheffield Partner Organisation (s) Tomsk State University (Russia); Hovd branch of Mongolian State University; Altai Botanical Gardens (Leninogorsk, Kazakhstan) Darwin Grant Value £184,316.84 Start/End dates 01.04.2002 – 31.03.2007 Reporting period and report 01.04.2005 – 31.03.2007 (Final report) number Project website http://www.ecos.tsu.ru/altai* Author(s), date Dr. -

Ventura County Plant Species of Local Concern
Checklist of Ventura County Rare Plants (Twenty-second Edition) CNPS, Rare Plant Program David L. Magney Checklist of Ventura County Rare Plants1 By David L. Magney California Native Plant Society, Rare Plant Program, Locally Rare Project Updated 4 January 2017 Ventura County is located in southern California, USA, along the east edge of the Pacific Ocean. The coastal portion occurs along the south and southwestern quarter of the County. Ventura County is bounded by Santa Barbara County on the west, Kern County on the north, Los Angeles County on the east, and the Pacific Ocean generally on the south (Figure 1, General Location Map of Ventura County). Ventura County extends north to 34.9014ºN latitude at the northwest corner of the County. The County extends westward at Rincon Creek to 119.47991ºW longitude, and eastward to 118.63233ºW longitude at the west end of the San Fernando Valley just north of Chatsworth Reservoir. The mainland portion of the County reaches southward to 34.04567ºN latitude between Solromar and Sequit Point west of Malibu. When including Anacapa and San Nicolas Islands, the southernmost extent of the County occurs at 33.21ºN latitude and the westernmost extent at 119.58ºW longitude, on the south side and west sides of San Nicolas Island, respectively. Ventura County occupies 480,996 hectares [ha] (1,188,562 acres [ac]) or 4,810 square kilometers [sq. km] (1,857 sq. miles [mi]), which includes Anacapa and San Nicolas Islands. The mainland portion of the county is 474,852 ha (1,173,380 ac), or 4,748 sq.