Predicting Toxicity from Gene Expression with Neural Networks
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Ibuprofen: Pharmacology, Therapeutics and Side Effects
Ibuprofen: Pharmacology, Therapeutics and Side Effects K.D. Rainsford Ibuprofen: Pharmacology, Therapeutics and Side Effects K.D. Rainsford Biomedical Research Centre Sheffield Hallam University Sheffield United Kingdom ISBN 978 3 0348 0495 0 ISBN 978 3 0348 0496 7 (eBook) DOI 10.1007/978 3 0348 0496 7 Springer Heidelberg New York Dordrecht London Library of Congress Control Number: 2012951702 # Springer Basel 2012 This work is subject to copyright. All rights are reserved by the Publisher, whether the whole or part of the material is concerned, specifically the rights of translation, reprinting, reuse of illustrations, recitation, broadcasting, reproduction on microfilms or in any other physical way, and transmission or information storage and retrieval, electronic adaptation, computer software, or by similar or dissimilar methodology now known or hereafter developed. Exempted from this legal reservation are brief excerpts in connection with reviews or scholarly analysis or material supplied specifically for the purpose of being entered and executed on a computer system, for exclusive use by the purchaser of the work. Duplication of this publication or parts thereof is permitted only under the provisions of the Copyright Law of the Publisher’s location, in its current version, and permission for use must always be obtained from Springer. Permissions for use may be obtained through RightsLink at the Copyright Clearance Center. Violations are liable to prosecution under the respective Copyright Law. The use of general descriptive names, registered names, trademarks, service marks, etc. in this publication does not imply, even in the absence of a specific statement, that such names are exempt from the relevant protective laws and regulations and therefore free for general use. -

Analysis of Synthetic Chemical Drugs in Adulterated Chinese Medicines by Capillary Electrophoresis/ Electrospray Ionization Mass Spectrometry
RAPID COMMUNICATIONS IN MASS SPECTROMETRY Rapid Commun. Mass Spectrom. 2001; 15: 1473±1480 Analysis of Synthetic chemical drugs in adulterated Chinese medicines by capillary electrophoresis/ electrospray ionization mass spectrometry Hsing Ling Cheng, Mei-Chun Tseng, Pei-Lun Tsai and Guor Rong Her* Department of Chemistry, National Taiwan University, Taipei, Taiwan, R.O.C. Received 20 April 2001; Revised 20 June 2001; Accepted 21 June 2001 Sixteen synthetic chemical drugs, often found in adulterated Chinese medicines, were studied by capillary electrophoresis/UV absorbance CE/UV)and capillary electrophoresis/electrospray ioniza- tion mass spectrometry CE/ESI-MS). Only nine peaks were detected with CZE/UV, but on-line CZE/ MS provided clear identification for most compounds. For a real sample of a Chinese medicinal preparation, a few adulterants were identified by their migration times and protonated molecular ions. For coeluting compounds, more reliable identification was achieved by MS/MS in selected reaction monitoring mode. Micellar electrokinetic chromatography MEKC)using sodium dodecyl sulfate SDS)provided better separation than capillary zone electrophoresis CZE),and, under optimal conditions, fourteen peaks were detected using UV detection. In ESI, the interference of SDS was less severe in positive ion mode than in negative ion mode. Up to 20 mM SDS could be used in direct coupling of MEKC with ESI-MS if the mass spectrometer was operated in positive ion mode. Because of better resolution in MEKC, adulterants can be identified without the use of MS/MS. Copyright # 2001 John Wiley & Sons, Ltd. The use of Chinese medicines to maintain human health and provide the main basis for identification,generally lack the to cure disease has a long and rich history. -

Computational Predictions of Glass-Forming Ability And
Article pubs.acs.org/molecularpharmaceutics Terms of Use Computational Predictions of Glass-Forming Ability and Crystallization Tendency of Drug Molecules † ‡ § † † Amjad Alhalaweh, Ahmad Alzghoul, Waseem Kaialy, Denny Mahlin, and Christel A. S. Bergström*, † Department of Pharmacy, Uppsala University, Uppsala Biomedical Centre, P.O. Box 580, SE-751 23 Uppsala, Sweden ‡ Department of Information Technology, Uppsala University, Lagerhyddsv.̈ 2, hus 1, Box 337, SE-751 05 Uppsala, Sweden § School of Pharmacy, Faculty of Science and Engineering, University of Wolverhampton, Wolverhampton WV1 1LY, United Kingdom ABSTRACT: Amorphization is an attractive formulation technique for drugs suffering from poor aqueous solubility as a result of their high lattice energy. Computational models that can predict the material properties associated with amorphiza- tion, such as glass-forming ability (GFA) and crystallization behavior in the dry state, would be a time-saving, cost-effective, and material-sparing approach compared to traditional experimental procedures. This article presents predictive models of these properties developed using support vector machine (SVM) algorithm. The GFA and crystallization tendency were investigated by melt-quenching 131 drug molecules in situ using differential scanning calorimetry. The SVM algorithm was used to develop computational models based on calculated molecular descriptors. The analyses confirmed the previously suggested cutoff molecular weight (MW) of 300 for glass-formers, and also clarified the extent to which MW can be used to predict the GFA of compounds with MW < 300. The topological equivalent of Grav3_3D, which is related to molecular size and shape, was a better descriptor than MW for GFA; it was able to accurately predict 86% of the data set regardless of MW. -

BMJ Open Is Committed to Open Peer Review. As Part of This Commitment We Make the Peer Review History of Every Article We Publish Publicly Available
BMJ Open is committed to open peer review. As part of this commitment we make the peer review history of every article we publish publicly available. When an article is published we post the peer reviewers’ comments and the authors’ responses online. We also post the versions of the paper that were used during peer review. These are the versions that the peer review comments apply to. The versions of the paper that follow are the versions that were submitted during the peer review process. They are not the versions of record or the final published versions. They should not be cited or distributed as the published version of this manuscript. BMJ Open is an open access journal and the full, final, typeset and author-corrected version of record of the manuscript is available on our site with no access controls, subscription charges or pay-per-view fees (http://bmjopen.bmj.com). If you have any questions on BMJ Open’s open peer review process please email [email protected] BMJ Open Pediatric drug utilization in the Western Pacific region: Australia, Japan, South Korea, Hong Kong and Taiwan Journal: BMJ Open ManuscriptFor ID peerbmjopen-2019-032426 review only Article Type: Research Date Submitted by the 27-Jun-2019 Author: Complete List of Authors: Brauer, Ruth; University College London, Research Department of Practice and Policy, School of Pharmacy Wong, Ian; University College London, Research Department of Practice and Policy, School of Pharmacy; University of Hong Kong, Centre for Safe Medication Practice and Research, Department -

WO 2013/020527 Al 14 February 2013 (14.02.2013) P O P C T
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization International Bureau (10) International Publication Number (43) International Publication Date WO 2013/020527 Al 14 February 2013 (14.02.2013) P O P C T (51) International Patent Classification: (74) Common Representative: UNIVERSITY OF VETER¬ A61K 9/06 (2006.01) A61K 47/32 (2006.01) INARY AND PHARMACEUTICAL SCIENCES A61K 9/14 (2006.01) A61K 47/38 (2006.01) BRNO FACULTY OF PHARMACY; University of A61K 47/10 (2006.01) A61K 9/00 (2006.01) Veterinary and Pharmaceutical Sciences Brno Faculty Of A61K 47/18 (2006.01) Pharmacy, Palackeho 1/3, CZ-61242 Brno (CZ). (21) International Application Number: (81) Designated States (unless otherwise indicated, for every PCT/CZ20 12/000073 kind of national protection available): AE, AG, AL, AM, AO, AT, AU, AZ, BA, BB, BG, BH, BN, BR, BW, BY, (22) Date: International Filing BZ, CA, CH, CL, CN, CO, CR, CU, CZ, DE, DK, DM, 2 August 2012 (02.08.2012) DO, DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, (25) Filing Language: English HN, HR, HU, ID, IL, IN, IS, JP, KE, KG, KM, KN, KP, KR, KZ, LA, LC, LK, LR, LS, LT, LU, LY, MA, MD, (26) Publication Language: English ME, MG, MK, MN, MW, MX, MY, MZ, NA, NG, NI, (30) Priority Data: NO, NZ, OM, PE, PG, PH, PL, PT, QA, RO, RS, RU, RW, 201 1-495 11 August 201 1 ( 11.08.201 1) SC, SD, SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, TM, 2012- 72 1 February 2012 (01.02.2012) TN, TR, TT, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, 2012-5 11 26 July 2012 (26.07.2012) ZW. -

Guidelines for Atcvet Classification 2021
Guidelines for ATCvet classification 2021 ISSN 1020-9891 ISBN 978-82-8406-167-2 Suggested citation: WHO Collaborating Centre for Drug Statistics Methodology, Guidelines for ATCvet classification 2021. Oslo, 2021. © Copyright WHO Collaborating Centre for Drug Statistics Methodology, Oslo, Norway. Use of all or parts of the material requires reference to the WHO Collaborating Centre for Drug Statistics Methodology. Copying and distribution for commercial purposes is not allowed. Changing or manipulating the material is not allowed. Guidelines for ATCvet classification 23rd edition WHO Collaborating Centre for Drug Statistics Methodology Norwegian Institute of Public Health P.O.Box 222 Skøyen N-0213 Oslo Norway Telephone: +47 21078160 E-mail: [email protected] Website: www.whocc.no Previous editions: 1992: Guidelines on ATCvet classification, 1st edition1) 1995: Guidelines on ATCvet classification, 2nd edition1) 1999: Guidelines on ATCvet classification, 3rd edition1) 2002: Guidelines for ATCvet classification, 4th edition2) 2003: Guidelines for ATCvet classification, 5th edition2) 2004: Guidelines for ATCvet classification, 6th edition2) 2005: Guidelines for ATCvet classification, 7th edition2) 2006: Guidelines for ATCvet classification, 8th edition2) 2007: Guidelines for ATCvet classification, 9th edition2) 2008: Guidelines for ATCvet classification, 10th edition2) 2009: Guidelines for ATCvet classification, 11th edition2) 2010: Guidelines for ATCvet classification, 12th edition2) 2011: Guidelines for ATCvet classification, 13th edition2) 2012: -

Page 1 S C Routine Toxicology Library Drug 3,4
Hospital for Sick Children Routine Toxicology Library of Drugs Detected Drug Drug 3,4-Methylenedioxyamphetamine Bupropion 3,4-Methylenedioxyethylamphetamine Buspirone 3,4-Methylenedioxymethamphetamine Butylone 6-O-Monoacetylmorphine Carbamazepine Acetaminophen Carbamazepine 10,11-epoxide Aciclovir Carbinoxamine Alprazolam Carvedilol α-Hydroxyalprazolam Cetirizine Amiloride Chlordiazepoxide Amiodarone Demoxepam Amitriptyline Chlorpheniramine Amlodipine Chlorpromazine Amoxapine Chlorprothixene Amoxicillin Cimetidine Amphetamine Ciprofloxacin Apomorphine Citalopram Aripiprazole Desmethylcitalopram Atenolol Clemastine Atorvastatin Clenbuterol Atropine Clobazam Baclofen Desmethylclobazam Benzatropine Clomethiazole Benzocaine Clomipramine Benzthiazide Desmethylclomipramine Benzylpiperazine Clonazepam Betaxolol 7-Aminoclonazepam Biperiden Clonidine Bisoprolol Clozapine Bromazepam N-Desmethyl Clozapine Bromocriptine Clozapine N-Oxide Brompheniramine Cocaine Bucetin Benzoylecgonine Bumetanide Ecgoninemethylester Bupivacaine Codeine Bupranolol Codeine-6-B-glucuronide Buprenorphine Coumatetralyl Buprenorphine Glucuronide Cyclobenzaprine Norbuprenorphine Desipramine Norbuprenorphine Glucuronide Dexamethasone Page 1 of 6 Hospital for Sick Children Routine Toxicology Library of Drugs Detected Drug Drug Dextromethorphan Fluoxetine Dextrorphan Norfluoxetine Diazepam Fluphenazine Nordiazepam Flurazepam 4-Hydroxynordiazepam Desalkylflurazepam Diclofenac 2-Hydroxyethylflurazepam Dihydrocodeine Fluvoxamine Dihydrocodeine-6-β-glucuronide Gabapentin Dihydroergotamine -

(12) Patent Application Publication (10) Pub
US 2004O156872A1 (19) United States (12) Patent Application Publication (10) Pub. No.: US 2004/0156872 A1 Bosch et al. (43) Pub. Date: Aug. 12, 2004 (54) NOVEL NMESULIDE COMPOSITIONS application No. 09/572,961, filed on May 18, 2000, now Pat. No. 6,316,029. (75) Inventors: H. William Bosch, Bryn Mawr, PA (US); Christian F. Wertz, Brookhaven, Publication Classification PA (US) (51) Int. Cl. ............................ A61K 9/14; A61K 9/00 Correspondence Address: FOLEY AND LARDNER (52) U.S. Cl. ............................................ 424/400; 424/489 SUTE 500 3000 KSTREET NW WASHINGTON, DC 20007 (US) (57) ABSTRACT (73) Assignee: Elan Pharma International Ltd. The present invention provides nanoparticulate nimeSulide (21) Appl. No.: 10/697,703 compositions. The compositions preferably comprise nime (22) Filed: Oct. 31, 2003 Sulide and at least one Surface Stabilizer adsorbed on or asSociated with the Surface of the nimeSulide particles. The Related U.S. Application Data nanoparticulate nimeSulide particles preferably have an effective average particle size of less than about 2000 nm. (63) Continuation-in-part of application No. 10/276,400, The invention also provides methods of making and using filed on Jan. 15, 2003, which is a continuation of nanoparticulate nimeSulide compositions. US 2004/O156872 A1 Aug. 12, 2004 NOVEL NMESULIDE COMPOSITIONS tion;' U.S. Pat. Nos. 5,399,363 and 5,494,683, both for “Surface Modified Anticancer Nanoparticles; U.S. Pat. No. RELATED APPLICATIONS 5,401,492 for “Water Insoluble Non-Magnetic Manganese Particles as Magnetic Resonance Enhancement Agents,” 0001. This application is a continuation-in-part of U.S. U.S. Pat. No. -

Pharmaceuticals (Monocomponent Products) ………………………..………… 31 Pharmaceuticals (Combination and Group Products) ………………….……
DESA The Department of Economic and Social Affairs of the United Nations Secretariat is a vital interface between global and policies in the economic, social and environmental spheres and national action. The Department works in three main interlinked areas: (i) it compiles, generates and analyses a wide range of economic, social and environmental data and information on which States Members of the United Nations draw to review common problems and to take stock of policy options; (ii) it facilitates the negotiations of Member States in many intergovernmental bodies on joint courses of action to address ongoing or emerging global challenges; and (iii) it advises interested Governments on the ways and means of translating policy frameworks developed in United Nations conferences and summits into programmes at the country level and, through technical assistance, helps build national capacities. Note Symbols of United Nations documents are composed of the capital letters combined with figures. Mention of such a symbol indicates a reference to a United Nations document. Applications for the right to reproduce this work or parts thereof are welcomed and should be sent to the Secretary, United Nations Publications Board, United Nations Headquarters, New York, NY 10017, United States of America. Governments and governmental institutions may reproduce this work or parts thereof without permission, but are requested to inform the United Nations of such reproduction. UNITED NATIONS PUBLICATION Copyright @ United Nations, 2005 All rights reserved TABLE OF CONTENTS Introduction …………………………………………………………..……..……..….. 4 Alphabetical Listing of products ……..………………………………..….….…..….... 8 Classified Listing of products ………………………………………………………… 20 List of codes for countries, territories and areas ………………………...…….……… 30 PART I. REGULATORY INFORMATION Pharmaceuticals (monocomponent products) ………………………..………… 31 Pharmaceuticals (combination and group products) ………………….……........ -

Practical Considerations for High-Throughput Chiral Screening in HPLC and SFC with 3- and Sub-2-Μm Particle-Packed Columns
14 May / June 2020 Practical Considerations for High-Throughput Chiral Screening in HPLC and SFC with 3- and Sub-2-µm Particle-Packed Columns Author: Edward G. Franklin Affiliation: Regis Technologies, Inc, 8210 Austin Ave, Morton Grove, IL 60053 The demands for increasingly fast chromatographic separations are unceasing as high-throughput methodologies are continuously sought and implemented in almost every analytical laboratory environment [1]. The pharmaceutical industry, for example, depends on fast analyses in order to speed the discovery and development of new drugs. Due to the complexity of biological targets, newly designed chemical entities often have one or more chiral centres, and, given certain FDA regulations, there is great need for fast assessment of enantiopurity and the feasibility of scaling for preparative purification. Chiral chromatography has long served as a reliable method for such analyses, but one of the major obstacles to the development of fast methods is the difficulty of predicting which chiral stationary phases (CSPs) will resolve the enantiomers of interest [2]. For this reason, samples are routinely screened against libraries of columns to determine which will perform best. This process often proceeds in a stepwise fashion and can be both manually intensive and time-consuming. To speed the process, labs have implemented automated, high-throughput screening approaches in order to identify promising CSPs before proceeding with further chromatographic method development. Such protocols may utilise instrumentation outfitted with 6- or 10- column selectors along with the ability to scout several mobile phases by incorporating quaternary pumping systems [3,4]. This approach also allows analysts to proceed with other tasks while chromatographic data is collected and processed over the course of a few hours or overnight. -
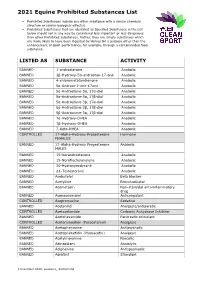
2021 Equine Prohibited Substances List
2021 Equine Prohibited Substances List . Prohibited Substances include any other substance with a similar chemical structure or similar biological effect(s). Prohibited Substances that are identified as Specified Substances in the List below should not in any way be considered less important or less dangerous than other Prohibited Substances. Rather, they are simply substances which are more likely to have been ingested by Horses for a purpose other than the enhancement of sport performance, for example, through a contaminated food substance. LISTED AS SUBSTANCE ACTIVITY BANNED 1-androsterone Anabolic BANNED 3β-Hydroxy-5α-androstan-17-one Anabolic BANNED 4-chlorometatandienone Anabolic BANNED 5α-Androst-2-ene-17one Anabolic BANNED 5α-Androstane-3α, 17α-diol Anabolic BANNED 5α-Androstane-3α, 17β-diol Anabolic BANNED 5α-Androstane-3β, 17α-diol Anabolic BANNED 5α-Androstane-3β, 17β-diol Anabolic BANNED 5β-Androstane-3α, 17β-diol Anabolic BANNED 7α-Hydroxy-DHEA Anabolic BANNED 7β-Hydroxy-DHEA Anabolic BANNED 7-Keto-DHEA Anabolic CONTROLLED 17-Alpha-Hydroxy Progesterone Hormone FEMALES BANNED 17-Alpha-Hydroxy Progesterone Anabolic MALES BANNED 19-Norandrosterone Anabolic BANNED 19-Noretiocholanolone Anabolic BANNED 20-Hydroxyecdysone Anabolic BANNED Δ1-Testosterone Anabolic BANNED Acebutolol Beta blocker BANNED Acefylline Bronchodilator BANNED Acemetacin Non-steroidal anti-inflammatory drug BANNED Acenocoumarol Anticoagulant CONTROLLED Acepromazine Sedative BANNED Acetanilid Analgesic/antipyretic CONTROLLED Acetazolamide Carbonic Anhydrase Inhibitor BANNED Acetohexamide Pancreatic stimulant CONTROLLED Acetominophen (Paracetamol) Analgesic BANNED Acetophenazine Antipsychotic BANNED Acetophenetidin (Phenacetin) Analgesic BANNED Acetylmorphine Narcotic BANNED Adinazolam Anxiolytic BANNED Adiphenine Antispasmodic BANNED Adrafinil Stimulant 1 December 2020, Lausanne, Switzerland 2021 Equine Prohibited Substances List . Prohibited Substances include any other substance with a similar chemical structure or similar biological effect(s). -

Harmonized Tariff Schedule of the United States (2004) -- Supplement 1 Annotated for Statistical Reporting Purposes
Harmonized Tariff Schedule of the United States (2004) -- Supplement 1 Annotated for Statistical Reporting Purposes PHARMACEUTICAL APPENDIX TO THE HARMONIZED TARIFF SCHEDULE Harmonized Tariff Schedule of the United States (2004) -- Supplement 1 Annotated for Statistical Reporting Purposes PHARMACEUTICAL APPENDIX TO THE TARIFF SCHEDULE 2 Table 1. This table enumerates products described by International Non-proprietary Names (INN) which shall be entered free of duty under general note 13 to the tariff schedule. The Chemical Abstracts Service (CAS) registry numbers also set forth in this table are included to assist in the identification of the products concerned. For purposes of the tariff schedule, any references to a product enumerated in this table includes such product by whatever name known. Product CAS No. Product CAS No. ABACAVIR 136470-78-5 ACEXAMIC ACID 57-08-9 ABAFUNGIN 129639-79-8 ACICLOVIR 59277-89-3 ABAMECTIN 65195-55-3 ACIFRAN 72420-38-3 ABANOQUIL 90402-40-7 ACIPIMOX 51037-30-0 ABARELIX 183552-38-7 ACITAZANOLAST 114607-46-4 ABCIXIMAB 143653-53-6 ACITEMATE 101197-99-3 ABECARNIL 111841-85-1 ACITRETIN 55079-83-9 ABIRATERONE 154229-19-3 ACIVICIN 42228-92-2 ABITESARTAN 137882-98-5 ACLANTATE 39633-62-0 ABLUKAST 96566-25-5 ACLARUBICIN 57576-44-0 ABUNIDAZOLE 91017-58-2 ACLATONIUM NAPADISILATE 55077-30-0 ACADESINE 2627-69-2 ACODAZOLE 79152-85-5 ACAMPROSATE 77337-76-9 ACONIAZIDE 13410-86-1 ACAPRAZINE 55485-20-6 ACOXATRINE 748-44-7 ACARBOSE 56180-94-0 ACREOZAST 123548-56-1 ACEBROCHOL 514-50-1 ACRIDOREX 47487-22-9 ACEBURIC ACID 26976-72-7