A Protocol for Constructing Gene Targeting Vectors: Generating Knockout Mice for the Cadherin Family and Beyond
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Molecular and Physiological Basis for Hair Loss in Near Naked Hairless and Oak Ridge Rhino-Like Mouse Models: Tracking the Role of the Hairless Gene
University of Tennessee, Knoxville TRACE: Tennessee Research and Creative Exchange Doctoral Dissertations Graduate School 5-2006 Molecular and Physiological Basis for Hair Loss in Near Naked Hairless and Oak Ridge Rhino-like Mouse Models: Tracking the Role of the Hairless Gene Yutao Liu University of Tennessee - Knoxville Follow this and additional works at: https://trace.tennessee.edu/utk_graddiss Part of the Life Sciences Commons Recommended Citation Liu, Yutao, "Molecular and Physiological Basis for Hair Loss in Near Naked Hairless and Oak Ridge Rhino- like Mouse Models: Tracking the Role of the Hairless Gene. " PhD diss., University of Tennessee, 2006. https://trace.tennessee.edu/utk_graddiss/1824 This Dissertation is brought to you for free and open access by the Graduate School at TRACE: Tennessee Research and Creative Exchange. It has been accepted for inclusion in Doctoral Dissertations by an authorized administrator of TRACE: Tennessee Research and Creative Exchange. For more information, please contact [email protected]. To the Graduate Council: I am submitting herewith a dissertation written by Yutao Liu entitled "Molecular and Physiological Basis for Hair Loss in Near Naked Hairless and Oak Ridge Rhino-like Mouse Models: Tracking the Role of the Hairless Gene." I have examined the final electronic copy of this dissertation for form and content and recommend that it be accepted in partial fulfillment of the requirements for the degree of Doctor of Philosophy, with a major in Life Sciences. Brynn H. Voy, Major Professor We have read this dissertation and recommend its acceptance: Naima Moustaid-Moussa, Yisong Wang, Rogert Hettich Accepted for the Council: Carolyn R. -

Supplementary Table 1: Adhesion Genes Data Set
Supplementary Table 1: Adhesion genes data set PROBE Entrez Gene ID Celera Gene ID Gene_Symbol Gene_Name 160832 1 hCG201364.3 A1BG alpha-1-B glycoprotein 223658 1 hCG201364.3 A1BG alpha-1-B glycoprotein 212988 102 hCG40040.3 ADAM10 ADAM metallopeptidase domain 10 133411 4185 hCG28232.2 ADAM11 ADAM metallopeptidase domain 11 110695 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 195222 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 165344 8751 hCG20021.3 ADAM15 ADAM metallopeptidase domain 15 (metargidin) 189065 6868 null ADAM17 ADAM metallopeptidase domain 17 (tumor necrosis factor, alpha, converting enzyme) 108119 8728 hCG15398.4 ADAM19 ADAM metallopeptidase domain 19 (meltrin beta) 117763 8748 hCG20675.3 ADAM20 ADAM metallopeptidase domain 20 126448 8747 hCG1785634.2 ADAM21 ADAM metallopeptidase domain 21 208981 8747 hCG1785634.2|hCG2042897 ADAM21 ADAM metallopeptidase domain 21 180903 53616 hCG17212.4 ADAM22 ADAM metallopeptidase domain 22 177272 8745 hCG1811623.1 ADAM23 ADAM metallopeptidase domain 23 102384 10863 hCG1818505.1 ADAM28 ADAM metallopeptidase domain 28 119968 11086 hCG1786734.2 ADAM29 ADAM metallopeptidase domain 29 205542 11085 hCG1997196.1 ADAM30 ADAM metallopeptidase domain 30 148417 80332 hCG39255.4 ADAM33 ADAM metallopeptidase domain 33 140492 8756 hCG1789002.2 ADAM7 ADAM metallopeptidase domain 7 122603 101 hCG1816947.1 ADAM8 ADAM metallopeptidase domain 8 183965 8754 hCG1996391 ADAM9 ADAM metallopeptidase domain 9 (meltrin gamma) 129974 27299 hCG15447.3 ADAMDEC1 ADAM-like, -

Learning from Cadherin Structures and Sequences: Affinity Determinants and Protein Architecture
Learning from cadherin structures and sequences: affinity determinants and protein architecture Klára Fels ıvályi Submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the Graduate School of Arts and Sciences COLUMBIA UNIVERSITY 2014 © 2014 Klara Felsovalyi All rights reserved ABSTRACT Learning from cadherin structures and sequences: affinity determinants and protein architecture Klara Felsovalyi Cadherins are a family of cell-surface proteins mediating adhesion that are important in development and maintenance of tissues. The family is defined by the repeating cadherin domain (EC) in their extracellular region, but they are diverse in terms of protein size, architecture and cellular function. The best-understood subfamily is the type I classical cadherins, which are found in vertebrates and have five EC domains. Among the five different type I classical cadherins, the binding interactions are highly specific in their homo- and heterophilic binding affinities, though their sequences are very similar. As previously shown, E- and N-cadherins, two prototypic members of the subfamily, differ in their homophilic K D by about an order of magnitude, while their heterophilic affinity is intermediate. To examine the source of the binding affinity differences among type I cadherins, we used crystal structures, analytical ultracentrifugation (AUC), surface plasmon resonance (SPR), and electron paramagnetic resonance (EPR) studies. Phylogenetic analysis and binding affinity behavior show that the type I cadherins can be further divided into two subgroups, with E- and N-cadherin representing each. In addition to the affinity differences in their wild-type binding through the strand-swapped interface, a second interface also shows an affinity difference between E- and N-cadherin. -

Acceleration in the DNA Methylation Age in Breast Cancer Tumours from Very Young Women Sara S
www.nature.com/scientificreports OPEN Acceleration in the DNA methylation age in breast cancer tumours from very young women Sara S. Oltra1, Maria Peña-Chilet1, Kirsty Flower2, María Teresa Martinez1, Elisa Alonso3, Octavio Burgues3, Ana Lluch1,4, James M. Flanagan 2 & Gloria Ribas1,4* Breast cancer in very young women (≤35 years; BCVY) presents more aggressive and complex biological features than their older counterparts (BCO). Our aim was to evaluate methylation diferences between BCVY and BCO and their DNA epigenetic age. EPIC and 450k Illumina methylation arrays were used in 67 breast cancer tumours, including 32 from BCVY, for methylation study and additionally we analysed their epigenetic age. We identifed 2 219 CpG sites diferently-methylated in BCVY vs. BCO (FDR < 0.05; β-value diference ± 0.1). The signature showed a general hypomethylation profle with a selective small hypermethylation profle located in open-sea regions in BCVY against BCO and normal tissue. Strikingly, BCVY presented a signifcant increased epigenetic age-acceleration compared with older women. The afected genes were enriched for pathways in neuronal-system pathways, cell communication, and matrix organisation. Validation in an independent sample highlighted consistent higher expression of HOXD9, and PCDH10 genes in BCVY. Regions implicated in the hypermethylation profle were involved in Notch signalling pathways, the immune system or DNA repair. We further validated HDAC5 expression in BCVY. We have identifed a DNA methylation signature that is specifc to BCVY and have shown that epigenetic age-acceleration is increased in BCVY. Breast cancer (BC) is the most common malignancy in women worldwide1. Approximately 6.6% of BCs are diag- nosed in women aged 40 or younger, and of all cancers diagnosed in this age group, 40% are BCs; the average risk of developing BC by age 40 is one in 1732 and unfortunately, they are not included in mammography screening programmes. -

Role and Regulation of the P53-Homolog P73 in the Transformation of Normal Human Fibroblasts
Role and regulation of the p53-homolog p73 in the transformation of normal human fibroblasts Dissertation zur Erlangung des naturwissenschaftlichen Doktorgrades der Bayerischen Julius-Maximilians-Universität Würzburg vorgelegt von Lars Hofmann aus Aschaffenburg Würzburg 2007 Eingereicht am Mitglieder der Promotionskommission: Vorsitzender: Prof. Dr. Dr. Martin J. Müller Gutachter: Prof. Dr. Michael P. Schön Gutachter : Prof. Dr. Georg Krohne Tag des Promotionskolloquiums: Doktorurkunde ausgehändigt am Erklärung Hiermit erkläre ich, dass ich die vorliegende Arbeit selbständig angefertigt und keine anderen als die angegebenen Hilfsmittel und Quellen verwendet habe. Diese Arbeit wurde weder in gleicher noch in ähnlicher Form in einem anderen Prüfungsverfahren vorgelegt. Ich habe früher, außer den mit dem Zulassungsgesuch urkundlichen Graden, keine weiteren akademischen Grade erworben und zu erwerben gesucht. Würzburg, Lars Hofmann Content SUMMARY ................................................................................................................ IV ZUSAMMENFASSUNG ............................................................................................. V 1. INTRODUCTION ................................................................................................. 1 1.1. Molecular basics of cancer .......................................................................................... 1 1.2. Early research on tumorigenesis ................................................................................. 3 1.3. Developing -

Detection of Somatic Variants from Genomic Data and Their Role in Neurodegenerative Diseases
Detection of somatic variants from genomic data and their role in neurodegenerative diseases Irene Lobón García Aquesta tesi doctoral està subjecta a la llicència Reconeixement 4.0. Espanya de Creative Commons. Esta tesis doctoral está sujeta a la licencia Reconocimiento 4.0. España de Creative Commons. This doctoral thesis is licensed under the Creative Commons Attribution 4.0. Spain License. Memoria presentada por Irene Lobón García para optar al grado de doctora por la Universidad de Barcelona Programa de Doctorado en Biomedicina Tesis realizada en el Instituto de Biología Evolutiva (CSIC-UPF) Detection oF somatic variants From genomic data and their role in neurodegenerative diseases Irene Lobón García Eduardo Soriano García Tomàs Marquès Bonet A mi Familia, “La paciencia es la madre de la ciencia” ReFranero español Acknowledgements De estos cinco años me llevo innumerables enseñanzas. Por supuesto muchas en lo profesional, pero incluso más en lo personal. Esta tesis ha sido un trabajo en grupo y sin el apoyo y ayuda de mucha gente hubiese sido imposible. En primer lugar, quiero agradecer a Eduardo su conFianza todos estos años y sobre todo el haberme introducido con sus proyectos en el tema que ahora me apasiona. También a Tomàs por aceptarme en su grupo como estudiante de máster y después hacer lo posible para que me quedase de alguna manera en el grupo. También por introducirme en el consorcio que ha sido fundamental para mi trabajo y porque nunca hubiese salido de mí pedir una estancia en Harvard. Esta experiencia solo ha sido posible gracias a vosotros. Los comienzos Fueron duros, después de tantos años de aprendizaje guiado el salto a la investigación es diFícil. -

Discovery of an O-Mannosylation Pathway Selectively Serving Cadherins and Protocadherins
Discovery of an O-mannosylation pathway selectively serving cadherins and protocadherins Ida Signe Bohse Larsena,1, Yoshiki Narimatsua,1, Hiren Jitendra Joshia,1, Lina Siukstaitea, Oliver J. Harrisonb, Julia Braschb, Kerry M. Goodmanb, Lars Hansena, Lawrence Shapirob,c,d, Barry Honigb,c,d,e, Sergey Y. Vakhrusheva, Henrik Clausena, and Adnan Halima,2,3 aDepartment of Cellular and Molecular Medicine, Faculty of Health Sciences, Copenhagen Center for Glycomics, University of Copenhagen, DK-2200 Copenhagen, Denmark; bDepartment of Biochemistry and Molecular Biophysics, Columbia University, New York, NY 10032; cZuckerman Mind Brain Behavior Institute, Columbia University, New York, NY 10032; dDepartment of Systems Biology, Columbia University, New York, NY 10032; and eHoward Hughes Medical Institute, Columbia University, New York, NY 10032 Edited by Stuart A. Kornfeld, Washington University School of Medicine, St. Louis, MO, and approved September 6, 2017 (received for review May 22, 2017) The cadherin (cdh) superfamily of adhesion molecules carry muscular dystrophies that have been designated α-dystroglyca- O-linked mannose (O-Man) glycans at highly conserved sites nopathies because deficient O-Man glycosylation of α-DG dis- localized to specific β-strands of their extracellular cdh (EC) domains. rupts the interaction between the dystrophin glycoprotein complex These O-Man glycans do not appear to be elongated like O-Man and the ECM (7–9). Several studies have also implicated de- glycans found on α-dystroglycan (α-DG), and we recently demon- ficiency of POMT2 with E-cdh dysfunction (10–12), although di- strated that initiation of cdh/protocadherin (pcdh) O-Man glycosyl- rect evidence for a role in glycosylation of cdhs and pcdhs is ation is not dependent on the evolutionary conserved POMT1/ missing. -

Downregulation of SNRPG Induces Cell Cycle Arrest and Sensitizes Human Glioblastoma Cells to Temozolomide by Targeting Myc Through a P53-Dependent Signaling Pathway
Cancer Biol Med 2020. doi: 10.20892/j.issn.2095-3941.2019.0164 ORIGINAL ARTICLE Downregulation of SNRPG induces cell cycle arrest and sensitizes human glioblastoma cells to temozolomide by targeting Myc through a p53-dependent signaling pathway Yulong Lan1,2*, Jiacheng Lou2*, Jiliang Hu1, Zhikuan Yu1, Wen Lyu1, Bo Zhang1,2 1Department of Neurosurgery, Shenzhen People’s Hospital, Second Clinical Medical College of Jinan University, The First Affiliated Hospital of Southern University of Science and Technology, Shenzhen 518020, China;2 Department of Neurosurgery, The Second Affiliated Hospital of Dalian Medical University, Dalian 116023, China ABSTRACT Objective: Temozolomide (TMZ) is commonly used for glioblastoma multiforme (GBM) chemotherapy. However, drug resistance limits its therapeutic effect in GBM treatment. RNA-binding proteins (RBPs) have vital roles in posttranscriptional events. While disturbance of RBP-RNA network activity is potentially associated with cancer development, the precise mechanisms are not fully known. The SNRPG gene, encoding small nuclear ribonucleoprotein polypeptide G, was recently found to be related to cancer incidence, but its exact function has yet to be elucidated. Methods: SNRPG knockdown was achieved via short hairpin RNAs. Gene expression profiling and Western blot analyses were used to identify potential glioma cell growth signaling pathways affected by SNRPG. Xenograft tumors were examined to determine the carcinogenic effects of SNRPG on glioma tissues. Results: The SNRPG-mediated inhibitory effect on glioma cells might be due to the targeted prevention of Myc and p53. In addition, the effects of SNRPG loss on p53 levels and cell cycle progression were found to be Myc-dependent. Furthermore, SNRPG was increased in TMZ-resistant GBM cells, and downregulation of SNRPG potentially sensitized resistant cells to TMZ, suggesting that SNRPG deficiency decreases the chemoresistance of GBM cells to TMZ via the p53 signaling pathway. -

13Q Deletion Syndrome Involving RB1: Characterization of a New Minimal Critical Region for Psychomotor Delay
G C A T T A C G G C A T genes Article 13q Deletion Syndrome Involving RB1: Characterization of a New Minimal Critical Region for Psychomotor Delay Flavia Privitera 1,2, Arianna Calonaci 3, Gabriella Doddato 1,2, Filomena Tiziana Papa 1,2, Margherita Baldassarri 1,2 , Anna Maria Pinto 4, Francesca Mari 1,2,4, Ilaria Longo 4, Mauro Caini 3, Daniela Galimberti 3, Theodora Hadjistilianou 5, Sonia De Francesco 5, Alessandra Renieri 1,2,4 and Francesca Ariani 1,2,4,* 1 Medical Genetics, University of Siena, 53100 Siena, Italy; fl[email protected] (F.P.); [email protected] (G.D.); fi[email protected] (F.T.P.); [email protected] (M.B.); [email protected] (F.M.); [email protected] (A.R.) 2 Med Biotech Hub and Competence Center, Department of Medical Biotechnologies, University of Siena, 53100 Siena, Italy 3 Unit of Pediatrics, Department of Maternal, Newborn and Child Health, Azienda Ospedaliera Universitaria Senese, Policlinico ‘Santa Maria alle Scotte’, 53100 Siena, Italy; [email protected] (A.C.); [email protected] (M.C.); [email protected] (D.G.) 4 Genetica Medica, Azienda Ospedaliera Universitaria Senese, 53100 Siena, Italy; [email protected] (A.M.P.); [email protected] (I.L.) 5 Unit of Ophthalmology and Retinoblastoma Referral Center, Department of Surgery, University of Siena, Policlinico ‘Santa Maria alle Scotte’, 53100 Siena, Italy; [email protected] (T.H.); [email protected] (S.D.F.) * Correspondence: [email protected]; Tel.: +39-057-723-3303 Citation: Privitera, F.; Calonaci, A.; Abstract: Retinoblastoma (RB) is an ocular tumor of the pediatric age caused by biallelic inactivation Doddato, G.; Papa, F.T.; Baldassarri, of the RB1 gene (13q14). -

1111111111111111111Inuuu11
1111111111111111111inuuu1111111111u~ (12) United States Patent (io) Patent No.: US 9,896,681 B2 Goodwin et al. (45) Date of Patent: *Feb. 20, 2018 (54) GENETIC REGULATION OF BONE AND 4,993,413 A 2/1991 McLeod CELLS BY ELECTROMAGNETIC 5,002,890 A 3/1991 Morrison STIMULATION FIELDS AND USES 5,026,650 A 6/1991 Schwarz 5,153,132 A 10/1992 Goodwin THEREOF 5,153,133 A 10/1992 Schwarz 5,155,034 A 10/1992 Wolf (71) Applicant: The United States of America as 5,155,035 A 10/1992 Schwarz Represented by the Administrator of 5,308,764 A 5/1994 Goodwin the National Aeronautics and Space 5,627,021 A 5/1997 Goodwin 5,846,807 A 12/1998 Goodwin Administration, Washington, DC (US) 6,485,963 B1 11/2002 Wolf 6,673,597 B2 1/2004 Wolf (72) Inventors: Thomas J. Goodwin, Kemah, TX 6,730,498 B1 5/2004 Goodwin (US); Linda C. Shackelford, Webster, 6,919,205 B2 7/2005 Brighton TX (US) 7,160,024 B2 1/2007 Dougherty, Sr. 7,179,217 B2 2/2007 Goodwin 7,456,019 B2 11/2008 Goodwin (73) Assignee: The United States of America as 2006/0229487 Al 10/2006 Goodwin represented by the National 2007/0105769 Al 5/2007 Simon Aeronautics and Space 2008/0138415 Al 6/2008 Hussain Administration, Washington, DC (US) 2009/0234417 Al 9/2009 Pastena 2011/0105959 Al 5/2011 OConnor (*) Notice: Subject to any disclaimer, the term of this patent is extended or adjusted under 35 OTHER PUBLICATIONS U.S.C. -
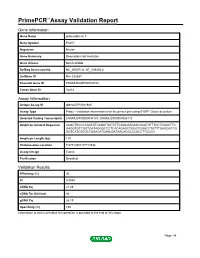
Primepcr™Assay Validation Report
PrimePCR™Assay Validation Report Gene Information Gene Name protocadherin 7 Gene Symbol Pcdh7 Organism Mouse Gene Summary Description Not Available Gene Aliases Not Available RefSeq Accession No. NC_000071.6, NT_039305.8 UniGene ID Mm.332387 Ensembl Gene ID ENSMUSG00000029108 Entrez Gene ID 54216 Assay Information Unique Assay ID qMmuCEP0041945 Assay Type Probe - Validation information is for the primer pair using SYBR® Green detection Detected Coding Transcript(s) ENSMUST00000094783, ENSMUST00000068110 Amplicon Context Sequence AAGCTGCCCCAATGTCAGATGATCTTCGACGAGAACGAATGTTTCCTGGACTTC GAGGTGTCGGTGATAGGGCCCTCACAGAGCTGGGTGGACCTGTTTGAGGGTCG GGTCATCGTGCTGGACATCAACGATAACACGCCCACCTTCCCG Amplicon Length (bp) 120 Chromosome Location 5:57719387-57719536 Assay Design Exonic Purification Desalted Validation Results Efficiency (%) 93 R2 0.9994 cDNA Cq 21.89 cDNA Tm (Celsius) 86 gDNA Cq 26.19 Specificity (%) 100 Information to assist with data interpretation is provided at the end of this report. Page 1/4 PrimePCR™Assay Validation Report Pcdh7, Mouse Amplification Plot Amplification of cDNA generated from 25 ng of universal reference RNA Melt Peak Melt curve analysis of above amplification Standard Curve Standard curve generated using 20 million copies of template diluted 10-fold to 20 copies Page 2/4 PrimePCR™Assay Validation Report Products used to generate validation data Real-Time PCR Instrument CFX384 Real-Time PCR Detection System Reverse Transcription Reagent iScript™ Advanced cDNA Synthesis Kit for RT-qPCR Real-Time PCR Supermix SsoAdvanced™ SYBR® Green Supermix Experimental Sample qPCR Mouse Reference Total RNA Data Interpretation Unique Assay ID This is a unique identifier that can be used to identify the assay in the literature and online. Detected Coding Transcript(s) This is a list of the Ensembl transcript ID(s) that this assay will detect. -

Supplementary Table 1 Double Treatment Vs Single Treatment
Supplementary table 1 Double treatment vs single treatment Probe ID Symbol Gene name P value Fold change TC0500007292.hg.1 NIM1K NIM1 serine/threonine protein kinase 1.05E-04 5.02 HTA2-neg-47424007_st NA NA 3.44E-03 4.11 HTA2-pos-3475282_st NA NA 3.30E-03 3.24 TC0X00007013.hg.1 MPC1L mitochondrial pyruvate carrier 1-like 5.22E-03 3.21 TC0200010447.hg.1 CASP8 caspase 8, apoptosis-related cysteine peptidase 3.54E-03 2.46 TC0400008390.hg.1 LRIT3 leucine-rich repeat, immunoglobulin-like and transmembrane domains 3 1.86E-03 2.41 TC1700011905.hg.1 DNAH17 dynein, axonemal, heavy chain 17 1.81E-04 2.40 TC0600012064.hg.1 GCM1 glial cells missing homolog 1 (Drosophila) 2.81E-03 2.39 TC0100015789.hg.1 POGZ Transcript Identified by AceView, Entrez Gene ID(s) 23126 3.64E-04 2.38 TC1300010039.hg.1 NEK5 NIMA-related kinase 5 3.39E-03 2.36 TC0900008222.hg.1 STX17 syntaxin 17 1.08E-03 2.29 TC1700012355.hg.1 KRBA2 KRAB-A domain containing 2 5.98E-03 2.28 HTA2-neg-47424044_st NA NA 5.94E-03 2.24 HTA2-neg-47424360_st NA NA 2.12E-03 2.22 TC0800010802.hg.1 C8orf89 chromosome 8 open reading frame 89 6.51E-04 2.20 TC1500010745.hg.1 POLR2M polymerase (RNA) II (DNA directed) polypeptide M 5.19E-03 2.20 TC1500007409.hg.1 GCNT3 glucosaminyl (N-acetyl) transferase 3, mucin type 6.48E-03 2.17 TC2200007132.hg.1 RFPL3 ret finger protein-like 3 5.91E-05 2.17 HTA2-neg-47424024_st NA NA 2.45E-03 2.16 TC0200010474.hg.1 KIAA2012 KIAA2012 5.20E-03 2.16 TC1100007216.hg.1 PRRG4 proline rich Gla (G-carboxyglutamic acid) 4 (transmembrane) 7.43E-03 2.15 TC0400012977.hg.1 SH3D19