Plant Genome Diversity Volume 2
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

The Down Rare Plant Register of Scarce & Threatened Vascular Plants
Vascular Plant Register County Down County Down Scarce, Rare & Extinct Vascular Plant Register and Checklist of Species Graham Day & Paul Hackney Record editor: Graham Day Authors of species accounts: Graham Day and Paul Hackney General editor: Julia Nunn 2008 These records have been selected from the database held by the Centre for Environmental Data and Recording at the Ulster Museum. The database comprises all known county Down records. The records that form the basis for this work were made by botanists, most of whom were amateur and some of whom were professional, employed by government departments or undertaking environmental impact assessments. This publication is intended to be of assistance to conservation and planning organisations and authorities, district and local councils and interested members of the public. Cover design by Fiona Maitland Cover photographs: Mourne Mountains from Murlough National Nature Reserve © Julia Nunn Hyoscyamus niger © Graham Day Spiranthes romanzoffiana © Graham Day Gentianella campestris © Graham Day MAGNI Publication no. 016 © National Museums & Galleries of Northern Ireland 1 Vascular Plant Register County Down 2 Vascular Plant Register County Down CONTENTS Preface 5 Introduction 7 Conservation legislation categories 7 The species accounts 10 Key to abbreviations used in the text and the records 11 Contact details 12 Acknowledgements 12 Species accounts for scarce, rare and extinct vascular plants 13 Casual species 161 Checklist of taxa from county Down 166 Publications relevant to the flora of county Down 180 Index 182 3 Vascular Plant Register County Down 4 Vascular Plant Register County Down PREFACE County Down is distinguished among Irish counties by its relatively diverse and interesting flora, as a consequence of its range of habitats and long coastline. -

Worksheet-2B.Pdf
WHAT’S SO IMPORTANT ABOUT NAMES? Topics Covered: Classificaon and taxonomy Understanding the importance of Linnaeus’s contribuon to science Making and using keys What’s in a name? Giving something a name allows us to talk about it. Names are important not only for people, but also for the plants we culvate in our gardens. In the early days of botany (the 17th and early 18th centuries) plants were given long Lan phrases for names that described their parcular features. As more plants became known, names tended to become longer, and much more difficult to remember and use. Then, in the 18th century, a Swedish biologist named Carl Linnaeus developed and popularised a two‐name (binomial) system for all plant species—GENUS and SPECIES. His system is sll in use today. A useful definion GENUS: A group of organisms SPECIES: that have certain characteriscs in of a species is a group of organisms common but can be divided further which can interbreed to produce into other groups (i.e. into species) ferle offspring Binomial names The use of only two words (the binomial name) made it much easier to categorise and compare different plants and animals. Imagine, for instance, talking about a type of geranium using the old name: Geranium pedunculis bifloris, caule dichotomo erecto, foliis quinquepars incisis; summis sessilibus The binomial name is much easier to use: Geranium maculatum 1 WHAT’S SO IMPORTANT ABOUT NAMES? Who was Carl Linnaeus? Carl Linnaeus (1707–1778) was born and brought up in and around Råshult, in the countryside of southern Sweden. -

Geranium Phaeum 'Joseph Green'
GERANIUM PHAEUM ‘JOSEPH GREEN’ Lynne Edwards y father, Joseph Green, a keen horticulturist, began his studies at Sutton Bonington School of M Agriculture in Nottinghamshire in 1937. His specialism was botany. He obtained the Certificate in Horticulture and an RHS Silver Medal. I have so much for which to thank him; he taught me much about botany as I was growing up, encouraging me to learn both the common and the botanical names of plants. As a teenager in the 1960s, right through to my father’s death in 2012, I gardened and Edwards All photographs © Lynne exchanged notes about flora with him. On his retirement, my father helped out at a friend’s plant nursery and, on one occasion, brought me a single-flowered, purple Geranium phaeum plant to fill an empty space in my garden. He warned me that if I allowed it to seed, there could be many new plants popping up all over the garden, adding that there might be the remote chance of a seedling producing a flower of an unusual colour and ‘how interesting that Geranium phaeum ‘Joseph Green’ would be!’ This turned out to be prophetic. Some years later, in early June 2013, I could not believe my eyes when I spotted a most unusual flowering plant in my garden, one I knew I had not planted. It was almost hidden between an old Rosa mutabilis and rampant brambles, and I could see from its leaves that it was a hardy geranium, yet its flowers of purple and chartreuse green were fascinating. It was growing in well-drained, light soil, amongst other G. -

An Encyclopedia of Shade Perennials This Page Intentionally Left Blank an Encyclopedia of Shade Perennials
An Encyclopedia of Shade Perennials This page intentionally left blank An Encyclopedia of Shade Perennials W. George Schmid Timber Press Portland • Cambridge All photographs are by the author unless otherwise noted. Copyright © 2002 by W. George Schmid. All rights reserved. Published in 2002 by Timber Press, Inc. Timber Press The Haseltine Building 2 Station Road 133 S.W. Second Avenue, Suite 450 Swavesey Portland, Oregon 97204, U.S.A. Cambridge CB4 5QJ, U.K. ISBN 0-88192-549-7 Printed in Hong Kong Library of Congress Cataloging-in-Publication Data Schmid, Wolfram George. An encyclopedia of shade perennials / W. George Schmid. p. cm. ISBN 0-88192-549-7 1. Perennials—Encyclopedias. 2. Shade-tolerant plants—Encyclopedias. I. Title. SB434 .S297 2002 635.9′32′03—dc21 2002020456 I dedicate this book to the greatest treasure in my life, my family: Hildegarde, my wife, friend, and supporter for over half a century, and my children, Michael, Henry, Hildegarde, Wilhelmina, and Siegfried, who with their mates have given us ten grandchildren whose eyes not only see but also appreciate nature’s riches. Their combined love and encouragement made this book possible. This page intentionally left blank Contents Foreword by Allan M. Armitage 9 Acknowledgments 10 Part 1. The Shady Garden 11 1. A Personal Outlook 13 2. Fated Shade 17 3. Practical Thoughts 27 4. Plants Assigned 45 Part 2. Perennials for the Shady Garden A–Z 55 Plant Sources 339 U.S. Department of Agriculture Hardiness Zone Map 342 Index of Plant Names 343 Color photographs follow page 176 7 This page intentionally left blank Foreword As I read George Schmid’s book, I am reminded that all gardeners are kindred in spirit and that— regardless of their roots or knowledge—the gardening they do and the gardens they create are always personal. -

Research on Spontaneous and Subspontaneous Flora of Botanical Garden "Vasile Fati" Jibou
Volume 19(2), 176- 189, 2015 JOURNAL of Horticulture, Forestry and Biotechnology www.journal-hfb.usab-tm.ro Research on spontaneous and subspontaneous flora of Botanical Garden "Vasile Fati" Jibou Szatmari P-M*.1,, Căprar M. 1 1) Biological Research Center, Botanical Garden “Vasile Fati” Jibou, Wesselényi Miklós Street, No. 16, 455200 Jibou, Romania; *Corresponding author. Email: [email protected] Abstract The research presented in this paper had the purpose of Key words inventory and knowledge of spontaneous and subspontaneous plant species of Botanical Garden "Vasile Fati" Jibou, Salaj, Romania. Following systematic Jibou Botanical Garden, investigations undertaken in the botanical garden a large number of spontaneous flora, spontaneous taxons were found from the Romanian flora (650 species of adventive and vascular plants and 20 species of moss). Also were inventoried 38 species of subspontaneous plants, adventive plants, permanently established in Romania and 176 vascular plant floristic analysis, Romania species that have migrated from culture and multiply by themselves throughout the garden. In the garden greenhouses were found 183 subspontaneous species and weeds, both from the Romanian flora as well as tropical plants introduced by accident. Thus the total number of wild species rises to 1055, a large number compared to the occupied area. Some rare spontaneous plants and endemic to the Romanian flora (Galium abaujense, Cephalaria radiata, Crocus banaticus) were found. Cultivated species that once migrated from culture, accommodated to environmental conditions and conquered new territories; standing out is the Cyrtomium falcatum fern, once escaped from the greenhouses it continues to develop on their outer walls. Jibou Botanical Garden is the second largest exotic species can adapt and breed further without any botanical garden in Romania, after "Anastasie Fătu" care [11]. -

1 Kedves Biológus Hallgató!
Biológiai és Ökológiai Intézet Természettudományi és Technológiai Kar Debreceni Egyetem 4032 Debrecen, Egyetem tér 1 Telefon: 52-316-666 Fax: 52-454-400 Kedves Biológus Hallgató! Köszöntünk a Debreceni Egyetem Természettudományi és Technológiai Karán. Szeretnénk, hogy sikeres és hasznos tagja légy az egyetemi polgárok nagy családjának és ezen belül is a szép hagyományokkal rendelkező biológusoknak. Kívánjuk, hogy nagyfokú érdeklődéssel és az új ismeretek befogadására nyitottan kezd el nálunk tanulmányaidat. Az Európai Felsőoktatási Térség kialakítását célzó – közismert nevén bolognai – folyamat megvalósításaként 2006. szeptemberétől a magyar felsőoktatásban is általánosan bevezetésre került a lineáris képzési rendszer, melynek szakaszai a következők: alap-(vagy BSc-) képzés 6 félév; mester-(vagy MSc-) képzés 4 félév; doktori (vagy PhD) képzés 6 félév. Ennek a nagyarányú átalakulásnak a keretében a Debreceni Egyetem Természettudományi és Technológiai Karán is elindultak az alapképzési szakok, melyek közül ez a kiadvány a Biológia alapszak tantervét és tantárgyi programjait tartalmazza. A könnyebb áttekinthetőség érdekében a tanterveket táblázatokban (tantervi hálók) is összefoglaltuk. Reméljük, hogy ez a füzet („fehér füzet”) segít majd Neked abban, hogy eligazodj az új közegben, és a felvehető tantárgyak széles választékából a legokosabban állítsd össze az órarendedet, hiszen ez meghatározza a következő évekre is a tanulmányaidat. A biológia alapképzést úgy terveztük meg, hogy a széles körű elméleti és gyakorlati ismeretekkel ruházza fel a végzettséget megszerzőket. Kérjük, ne feledd, hogy a tudást nem adják ingyen, azért meg kell dolgozni. Ebben a munkában a biológus és más szakmabeli oktatók, illetve egyéb dolgozók a partnereid lesznek, együttműködésükre mindig számíthatsz. Bízunk benne, hogy ennek az együttes munkának a gyümölcse egy jó elhelyezkedési lehetőségeket biztosító diploma, illetve a mesterképzésbe való továbblépés lesz. -
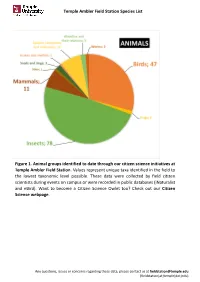
Temple Ambler Field Station Species List Figure 1. Animal Groups Identified to Date Through Our Citizen Science Initiatives at T
Temple Ambler Field Station Species List Figure 1. Animal groups identified to date through our citizen science initiatives at Temple Ambler Field Station. Values represent unique taxa identified in the field to the lowest taxonomic level possible. These data were collected by field citizen scientists during events on campus or were recorded in public databases (iNaturalist and eBird). Want to become a Citizen Science Owlet too? Check out our Citizen Science webpage. Any questions, issues or concerns regarding these data, please contact us at [email protected] (fieldstation[at}temple[dot]edu) Temple Ambler Field Station Species List Figure 2. Plant diversity identified to date in the natural environments and designed gardens of the Temple Ambler Field Station and Ambler Arboretum. These values represent unique taxa identified to the lowest taxonomic level possible. Highlighted are 14 of the 116 flowering plant families present that include 524 taxonomic groups. A full list can be found in our species database. Cultivated specimens in our Greenhouse were not included here. Any questions, issues or concerns regarding these data, please contact us at [email protected] (fieldstation[at}temple[dot]edu) Temple Ambler Field Station Species List database_title Temple Ambler Field Station Species List last_update 22October2020 description This database includes all species identified to their lowest taxonomic level possible in the natural environments and designed gardens on the Temple Ambler campus. These are occurrence records and each taxon is only entered once. This is an occurrence record, not an abundance record. IDs were performed by senior scientists and specialists, as well as citizen scientists visiting campus. -

Samenkatalog Graz 2019 End.Pdf
SAMENTAUSCHVERZEICHNIS Index Seminum Seed list Catalogue de graines Botanischer Garten der Karl-Franzens-Universität Graz Ernte / Harvest / Récolte 2019 Herausgegeben von Christian BERG, Kurt MARQUART, Thomas GALIK & Jonathan WILFLING ebgconsortiumindexseminum2012 Institut für Biology, Januar 2020 Botanical Garden, Institute of Biology, Karl-Franzens-Universität Graz 2 Botanischer Garten Institut für Biologie Karl-Franzens-Universität Graz Holteigasse 6 A - 8010 Graz, Austria Fax: ++43-316-380-9883 Email- und Telefonkontakt: [email protected], Tel.: ++43-316-380-5651 [email protected], Tel.: ++43-316-380-5747 Webseite: http://garten.uni-graz.at/ Zitiervorschlag : BERG, C., MARQUART, K., GALIK, T. & Wilfling, J. (2020): Samentauschverzeichnis – Index Seminum – des Botanischen Gartens der Karl-Franzens-Universität Graz, Samenernte 2019. – 44 S., Karl-Franzens-Universität Graz. Personalstand des Botanischen Gartens Graz: Institutsleiter: Univ.-Prof. Dr. Christian STURMBAUER Wissenschaftlicher Gartenleiter: Dr. Christian BERG Technischer Gartenleiter: Jonathan WILFLING, B. Sc. GärtnerInnen: Doris ADAM-LACKNER Viola BONGERS Thomas GALIK Margarete HIDEN Kurt MARQUART Franz STIEBER Ulrike STRAUSSBERGER Monika GABER René MICHALSKI Techn. MitarbeiterInnen: Oliver KROPIWNICKI Martina THALHAMMER Gärtnerlehrlinge: Sophia DAMBRICH (3. Lehrjahr) Wanja WIRTL-MÖLBACH (3. Lehrjahr) Jean KERSCHBAUMER (3. Lehrjahr) 3 Inhaltsverzeichnis / Contents / Table des matières Abkürzungen / List of abbreviations / Abréviations ................................................. -

Home Orchard Home Orchard
TheThe AmericanAmerican GARDENERGARDENER® TheThe MagazineMagazine ofof thethe AAmericanmerican HorticulturalHorticultural SocietySociety May / June 2008 rewards of a Home Orchard Impatiens: Beyond Busy Lizzies A Love for Lilies Hardy Geraniums for Carefree Color Your plant’s nutritionist. (Actual size) Introducing Osmocote® Plus Multi- Purpose Plant Food. All the nutrition your plants need is packed into this tiny granule. New Osmocote® Plus is formulated with the 12 nutrients that help plants flourish and a Smart-Release® coating that knows exactly when to feed. It nourishes up to six full months, won’t wash away and won’t burn. Now, no matter what you grow, or where you grow it, Osmocote® Plus is the only plant food you’ll need. Let’s just say, we’ve got plant nutrition down to an exact science. It knows. PlantersPlace.com/Plus © 2008 The Scotts Company LLC. World rights reserved. contents Volume 87, Number 3 . May / June 2008 FEATURES DEPARTMENTS 5 NOTES FROM RIVER FARM 6 MEMBERS’ FORUM 7 NEWS FROM AHS Green Garage is part of U.S. Botanic Garden exhibition, Garden School on trees a success, winners of the AHS Environmental Award at Philadelphia and San Francisco flower shows, upcoming webinars on designing with color and woodland gardens. 12 AHS NEWS SPECIAL: 2008 NATIONAL CHILDREN & YOUTH GARDEN SYMPOSIUM An overview of what page 27 is in store for this year’s event in the Philadelphia area. 16 CAREFREE CRANESBILLS BY RICHARD HAWKE 44 ONE ON ONE WITH… With their long blooming season and easy culture, hardy geraniums Harold Pellett, plant page 12 are tough to beat. -

Contrasting Patterns of Nucleotide Substitution Rates Provide Insight Into Dynamic Evolution of Plastid and Mitochondrial Genomes of Geranium
GBE Contrasting Patterns of Nucleotide Substitution Rates Provide Insight into Dynamic Evolution of Plastid and Mitochondrial Genomes of Geranium Seongjun Park1,TraceyA.Ruhlman1,Mao-LunWeng1,2, Nahid H. Hajrah3, Jamal S.M. Sabir3, and Robert K. Jansen1,3,* 1Department of Integrative Biology, University of Texas at Austin 2Department of Biology and Microbiology, South Dakota State University 3Genomic and Biotechnology Research Group, Department of Biological Science, Faculty of Science, King Abdulaziz University, Jeddah, Saudi Arabia *Corresponding author: E-mail: [email protected]. Accepted: July 3, 2017 Data deposition: All sequences used in this study have been submitted to NCBI Genbank and accession numbers are reported in supplementary tables S10–12, Supplementary Material online. Abstract Geraniaceae have emerged as a model system for investigating the causes and consequences of variation in plastid and mitochon- drial genomes. Incredible structural variation in plastid genomes (plastomes) and highly accelerated evolutionary rates have been reported in selected lineages and functional groups of genes in both plastomes and mitochondrial genomes (mitogenomes), and these phenomena have been implicated in cytonuclear incompatibility. Previous organelle genome studies have included limited sampling of Geranium, the largest genus in the family with over 400 species. This study reports on rates and patterns of nucleotide substitutions in plastomes and mitogenomes of 17 species of Geranium and representatives of other Geraniaceae. As detected across other angiosperms, substitution rates in the plastome are 3.5 times higher than the mitogenome in most Geranium. However, in the branch leading to Geranium brycei/Geranium incanum mitochondrial genes experienced signifi- cantly higher dN and dS than plastid genes, a pattern that has only been detected in one other angiosperm. -
Plastid Genome Evolution ADVANCES in BOTANICAL RESEARCH
VOLUME EIGHTY FIVE ADVANCES IN BOTANICAL RESEARCH Plastid Genome Evolution ADVANCES IN BOTANICAL RESEARCH Series Editors Jean-Pierre Jacquot Professor, Membre de L’Institut Universitaire de France, Unite´ Mixte de Recherche INRA, UHP 1136 “Interaction Arbres Microorganismes”, Universite de Lorraine, Faculte des Sciences, Vandoeuvre, France Pierre Gadal Honorary Professor, Universite Paris-Sud XI, Institut Biologie des Plantes, Orsay, France VOLUME EIGHTY FIVE ADVANCES IN BOTANICAL RESEARCH Plastid Genome Evolution Volume Editors SHU-MIAW CHAW Biodiversity Research Center, Academia Sinica; Biodiversity Program, Taiwan International Graduate Program, Academia Sinica and National Taiwan Normal University, Taipei, Taiwan ROBERT K. JANSEN Department of Integrative Biology, University of Texas at Austin, Austin, TX, United States; Genomics and Biotechnology Research Group, Faculty of Science, King Abdulaziz University, Jeddah, Saudi Arabia Academic Press is an imprint of Elsevier 125 London Wall, London, EC2Y 5AS, United Kingdom The Boulevard, Langford Lane, Kidlington, Oxford OX5 1GB, United Kingdom 50 Hampshire Street, 5th Floor, Cambridge, MA 02139, United States 525 B Street, Suite 1800, San Diego, CA 92101-4495, United States First edition 2018 Copyright © 2018 Elsevier Ltd. All rights reserved. No part of this publication may be reproduced or transmitted in any form or by any means, electronic or mechanical, including photocopying, recording, or any information storage and retrieval system, without permission in writing from the publisher. Details on how to seek permission, further information about the Publisher’s permissions policies and our arrangements with organizations such as the Copyright Clearance Center and the Copyright Licensing Agency, can be found at our website: www.elsevier.com/permissions. This book and the individual contributions contained in it are protected under copyright by the Publisher (other than as may be noted herein). -

An Ecological Database of the British Flora
An Ecological Database of the British Flora submitted by Helen Jacqueline Peat for examination for the degree Doctor of Philosophy Department of Biology University of York October 1992 Abstract The design and compilation of a database containing ecological information on the British Flora is described. All native and naturalised species of the Gymnospermae and Angiospermae are included. Data on c.130 characteristics concerning habitat, distribution, morphology, physiology, life history and associated organisms, were collected by both literature searching and correspondence with plant ecologists. The evolutionary history of 25 of the characteristics was investigated by looking at the amount of variance at each taxonomic level. The variation in pollination mechanisms was found at high taxonomic levels suggesting these evolved, and became fixed, early on in the evolution of flowering plants. Chromosome number, annualness, dichogamy and self-fertilization showed most variance at low taxonomic levels, suggesting these characteristics have evolved more recently and may still be subject to change. Most of the characteristics, however, eg. presence of compound leaves, height and propagule length showed variance spread over several taxonomic levels suggesting evolution has occurred at different times in different lineages. The necessity of accounting for phylogeny when conducting comparative analyses is discussed, and two methods allowing this are outlined. Using these, the questions: 'Why does stomatal distribution differ between species?' and 'Why do different species have different degrees of mycorrhizal infection?' were investigated. Amphistomaty was found to be associated with species of unshaded habitats, those with small leaves and those with hairy leaves, and hypostomaty with woody species, larger leaves and glabrous leaves.