Genetics and Genomics of Mammalian Pigment Patterns
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
PLAGUE STUDIES * 6. Hosts of the Infection R
Bull. Org. mond. Sante 1 Bull. World Hlth Org. 1952, 6, 381-465 PLAGUE STUDIES * 6. Hosts of the Infection R. POLLITZER, M.D. Division of Epidemiology, World Health Organization Manuscript received in April 1952 RODENTS AND LAGOMORPHA Reviewing in 1928 the then rather limited knowledge available concerning the occurrence and importance of plague in rodents other than the common rats and mice, Jorge 129 felt justified in drawing a clear-cut distinction between the pandemic type of plague introduced into human settlements and houses all over the world by the " domestic " rats and mice, and " peste selvatique ", which is dangerous for man only when he invades the remote endemic foci populated by wild rodents. Although Jorge's concept was accepted, some discussion arose regarding the appropriateness of the term " peste selvatique" or, as Stallybrass 282 and Wu Lien-teh 318 translated it, " selvatic plague ". It was pointed out by Meyer 194 that, on etymological grounds, the name " sylvatic plague " would be preferable, and this term was widely used until POzzO 238 and Hoekenga 105 doubted, and Girard 82 denied, its adequacy on the grounds that the word " sylvatic" implied that the rodents concerned lived in forests, whereas that was rarely the case. Girard therefore advocated the reversion to the expression "wild-rodent plague" which was used before the publication of Jorge's study-a proposal it has seemed advisable to accept for the present studies. Much more important than the difficulty of adopting an adequate nomenclature is that of distinguishing between rat and wild-rodent plague- a distinction which is no longer as clear-cut as Jorge was entitled to assume. -

Species List
Mozambique: Species List Birds Specie Seen Location Common Quail Harlequin Quail Blue Quail Helmeted Guineafowl Crested Guineafowl Fulvous Whistling-Duck White-faced Whistling-Duck White-backed Duck Egyptian Goose Spur-winged Goose Comb Duck African Pygmy-Goose Cape Teal African Black Duck Yellow-billed Duck Cape Shoveler Red-billed Duck Northern Pintail Hottentot Teal Southern Pochard Small Buttonquail Black-rumped Buttonquail Scaly-throated Honeyguide Greater Honeyguide Lesser Honeyguide Pallid Honeyguide Green-backed Honeyguide Wahlberg's Honeyguide Rufous-necked Wryneck Bennett's Woodpecker Reichenow's Woodpecker Golden-tailed Woodpecker Green-backed Woodpecker Cardinal Woodpecker Stierling's Woodpecker Bearded Woodpecker Olive Woodpecker White-eared Barbet Whyte's Barbet Green Barbet Green Tinkerbird Yellow-rumped Tinkerbird Yellow-fronted Tinkerbird Red-fronted Tinkerbird Pied Barbet Black-collared Barbet Brown-breasted Barbet Crested Barbet Red-billed Hornbill Southern Yellow-billed Hornbill Crowned Hornbill African Grey Hornbill Pale-billed Hornbill Trumpeter Hornbill Silvery-cheeked Hornbill Southern Ground-Hornbill Eurasian Hoopoe African Hoopoe Green Woodhoopoe Violet Woodhoopoe Common Scimitar-bill Narina Trogon Bar-tailed Trogon European Roller Lilac-breasted Roller Racket-tailed Roller Rufous-crowned Roller Broad-billed Roller Half-collared Kingfisher Malachite Kingfisher African Pygmy-Kingfisher Grey-headed Kingfisher Woodland Kingfisher Mangrove Kingfisher Brown-hooded Kingfisher Striped Kingfisher Giant Kingfisher Pied -

Identification of the Binding Partners for Hspb2 and Cryab Reveals
Brigham Young University BYU ScholarsArchive Theses and Dissertations 2013-12-12 Identification of the Binding arP tners for HspB2 and CryAB Reveals Myofibril and Mitochondrial Protein Interactions and Non- Redundant Roles for Small Heat Shock Proteins Kelsey Murphey Langston Brigham Young University - Provo Follow this and additional works at: https://scholarsarchive.byu.edu/etd Part of the Microbiology Commons BYU ScholarsArchive Citation Langston, Kelsey Murphey, "Identification of the Binding Partners for HspB2 and CryAB Reveals Myofibril and Mitochondrial Protein Interactions and Non-Redundant Roles for Small Heat Shock Proteins" (2013). Theses and Dissertations. 3822. https://scholarsarchive.byu.edu/etd/3822 This Thesis is brought to you for free and open access by BYU ScholarsArchive. It has been accepted for inclusion in Theses and Dissertations by an authorized administrator of BYU ScholarsArchive. For more information, please contact [email protected], [email protected]. Identification of the Binding Partners for HspB2 and CryAB Reveals Myofibril and Mitochondrial Protein Interactions and Non-Redundant Roles for Small Heat Shock Proteins Kelsey Langston A thesis submitted to the faculty of Brigham Young University in partial fulfillment of the requirements for the degree of Master of Science Julianne H. Grose, Chair William R. McCleary Brian Poole Department of Microbiology and Molecular Biology Brigham Young University December 2013 Copyright © 2013 Kelsey Langston All Rights Reserved ABSTRACT Identification of the Binding Partners for HspB2 and CryAB Reveals Myofibril and Mitochondrial Protein Interactors and Non-Redundant Roles for Small Heat Shock Proteins Kelsey Langston Department of Microbiology and Molecular Biology, BYU Master of Science Small Heat Shock Proteins (sHSP) are molecular chaperones that play protective roles in cell survival and have been shown to possess chaperone activity. -

Supplementary Information Integrative Analyses of Splicing in the Aging Brain: Role in Susceptibility to Alzheimer’S Disease
Supplementary Information Integrative analyses of splicing in the aging brain: role in susceptibility to Alzheimer’s Disease Contents 1. Supplementary Notes 1.1. Religious Orders Study and Memory and Aging Project 1.2. Mount Sinai Brain Bank Alzheimer’s Disease 1.3. CommonMind Consortium 1.4. Data Availability 2. Supplementary Tables 3. Supplementary Figures Note: Supplementary Tables are provided as separate Excel files. 1. Supplementary Notes 1.1. Religious Orders Study and Memory and Aging Project Gene expression data1. Gene expression data were generated using RNA- sequencing from Dorsolateral Prefrontal Cortex (DLPFC) of 540 individuals, at an average sequence depth of 90M reads. Detailed description of data generation and processing was previously described2 (Mostafavi, Gaiteri et al., under review). Samples were submitted to the Broad Institute’s Genomics Platform for transcriptome analysis following the dUTP protocol with Poly(A) selection developed by Levin and colleagues3. All samples were chosen to pass two initial quality filters: RNA integrity (RIN) score >5 and quantity threshold of 5 ug (and were selected from a larger set of 724 samples). Sequencing was performed on the Illumina HiSeq with 101bp paired-end reads and achieved coverage of 150M reads of the first 12 samples. These 12 samples will serve as a deep coverage reference and included 2 males and 2 females of nonimpaired, mild cognitive impaired, and Alzheimer's cases. The remaining samples were sequenced with target coverage of 50M reads; the mean coverage for the samples passing QC is 95 million reads (median 90 million reads). The libraries were constructed and pooled according to the RIN scores such that similar RIN scores would be pooled together. -

Open Dogan Phdthesis Final.Pdf
The Pennsylvania State University The Graduate School Eberly College of Science ELUCIDATING BIOLOGICAL FUNCTION OF GENOMIC DNA WITH ROBUST SIGNALS OF BIOCHEMICAL ACTIVITY: INTEGRATIVE GENOME-WIDE STUDIES OF ENHANCERS A Dissertation in Biochemistry, Microbiology and Molecular Biology by Nergiz Dogan © 2014 Nergiz Dogan Submitted in Partial Fulfillment of the Requirements for the Degree of Doctor of Philosophy August 2014 ii The dissertation of Nergiz Dogan was reviewed and approved* by the following: Ross C. Hardison T. Ming Chu Professor of Biochemistry and Molecular Biology Dissertation Advisor Chair of Committee David S. Gilmour Professor of Molecular and Cell Biology Anton Nekrutenko Professor of Biochemistry and Molecular Biology Robert F. Paulson Professor of Veterinary and Biomedical Sciences Philip Reno Assistant Professor of Antropology Scott B. Selleck Professor and Head of the Department of Biochemistry and Molecular Biology *Signatures are on file in the Graduate School iii ABSTRACT Genome-wide measurements of epigenetic features such as histone modifications, occupancy by transcription factors and coactivators provide the opportunity to understand more globally how genes are regulated. While much effort is being put into integrating the marks from various combinations of features, the contribution of each feature to accuracy of enhancer prediction is not known. We began with predictions of 4,915 candidate erythroid enhancers based on genomic occupancy by TAL1, a key hematopoietic transcription factor that is strongly associated with gene induction in erythroid cells. Seventy of these DNA segments occupied by TAL1 (TAL1 OSs) were tested by transient transfections of cultured hematopoietic cells, and 56% of these were active as enhancers. Sixty-six TAL1 OSs were evaluated in transgenic mouse embryos, and 65% of these were active enhancers in various tissues. -

Ingenuity Pathway Analysis of Differentially Expressed Genes Involved in Signaling Pathways and Molecular Networks in Rhoe Gene‑Edited Cardiomyocytes
INTERNATIONAL JOURNAL OF MOleCular meDICine 46: 1225-1238, 2020 Ingenuity pathway analysis of differentially expressed genes involved in signaling pathways and molecular networks in RhoE gene‑edited cardiomyocytes ZHONGMING SHAO1*, KEKE WANG1*, SHUYA ZHANG2, JIANLING YUAN1, XIAOMING LIAO1, CAIXIA WU1, YUAN ZOU1, YANPING HA1, ZHIHUA SHEN1, JUNLI GUO2 and WEI JIE1,2 1Department of Pathology, School of Basic Medicine Sciences, Guangdong Medical University, Zhanjiang, Guangdong 524023; 2Hainan Provincial Key Laboratory for Tropical Cardiovascular Diseases Research and Key Laboratory of Emergency and Trauma of Ministry of Education, Institute of Cardiovascular Research of The First Affiliated Hospital, Hainan Medical University, Haikou, Hainan 571199, P.R. China Received January 7, 2020; Accepted May 20, 2020 DOI: 10.3892/ijmm.2020.4661 Abstract. RhoE/Rnd3 is an atypical member of the Rho super- injury and abnormalities, cell‑to‑cell signaling and interaction, family of proteins, However, the global biological function and molecular transport. In addition, 885 upstream regulators profile of this protein remains unsolved. In the present study, a were enriched, including 59 molecules that were predicated RhoE‑knockout H9C2 cardiomyocyte cell line was established to be strongly activated (Z‑score >2) and 60 molecules that using CRISPR/Cas9 technology, following which differentially were predicated to be significantly inhibited (Z‑scores <‑2). In expressed genes (DEGs) between the knockout and wild‑type particular, 33 regulatory effects and 25 networks were revealed cell lines were screened using whole genome expression gene to be associated with the DEGs. Among them, the most signifi- chips. A total of 829 DEGs, including 417 upregulated and cant regulatory effects were ‘adhesion of endothelial cells’ and 412 downregulated, were identified using the threshold of ‘recruitment of myeloid cells’ and the top network was ‘neuro- fold changes ≥1.2 and P<0.05. -

Protein Kinase A-Mediated Septin7 Phosphorylation Disrupts Septin Filaments and Ciliogenesis
cells Article Protein Kinase A-Mediated Septin7 Phosphorylation Disrupts Septin Filaments and Ciliogenesis Han-Yu Wang 1,2, Chun-Hsiang Lin 1, Yi-Ru Shen 1, Ting-Yu Chen 2,3, Chia-Yih Wang 2,3,* and Pao-Lin Kuo 1,2,4,* 1 Department of Obstetrics and Gynecology, College of Medicine, National Cheng Kung University, Tainan 701, Taiwan; [email protected] (H.-Y.W.); [email protected] (C.-H.L.); [email protected] (Y.-R.S.) 2 Institute of Basic Medical Sciences, College of Medicine, National Cheng Kung University, Tainan 701, Taiwan; [email protected] 3 Department of Cell Biology and Anatomy, College of Medicine, National Cheng Kung University, Tainan 701, Taiwan 4 Department of Obstetrics and Gynecology, National Cheng-Kung University Hospital, Tainan 704, Taiwan * Correspondence: [email protected] (C.-Y.W.); [email protected] (P.-L.K.); Tel.: +886-6-2353535 (ext. 5338); (C.-Y.W.)+886-6-2353535 (ext. 5262) (P.-L.K.) Abstract: Septins are GTP-binding proteins that form heteromeric filaments for proper cell growth and migration. Among the septins, septin7 (SEPT7) is an important component of all septin filaments. Here we show that protein kinase A (PKA) phosphorylates SEPT7 at Thr197, thus disrupting septin filament dynamics and ciliogenesis. The Thr197 residue of SEPT7, a PKA phosphorylating site, was conserved among different species. Treatment with cAMP or overexpression of PKA catalytic subunit (PKACA2) induced SEPT7 phosphorylation, followed by disruption of septin filament formation. Constitutive phosphorylation of SEPT7 at Thr197 reduced SEPT7-SEPT7 interaction, but did not affect SEPT7-SEPT6-SEPT2 or SEPT4 interaction. -

1 Supporting Information for a Microrna Network Regulates
Supporting Information for A microRNA Network Regulates Expression and Biosynthesis of CFTR and CFTR-ΔF508 Shyam Ramachandrana,b, Philip H. Karpc, Peng Jiangc, Lynda S. Ostedgaardc, Amy E. Walza, John T. Fishere, Shaf Keshavjeeh, Kim A. Lennoxi, Ashley M. Jacobii, Scott D. Rosei, Mark A. Behlkei, Michael J. Welshb,c,d,g, Yi Xingb,c,f, Paul B. McCray Jr.a,b,c Author Affiliations: Department of Pediatricsa, Interdisciplinary Program in Geneticsb, Departments of Internal Medicinec, Molecular Physiology and Biophysicsd, Anatomy and Cell Biologye, Biomedical Engineeringf, Howard Hughes Medical Instituteg, Carver College of Medicine, University of Iowa, Iowa City, IA-52242 Division of Thoracic Surgeryh, Toronto General Hospital, University Health Network, University of Toronto, Toronto, Canada-M5G 2C4 Integrated DNA Technologiesi, Coralville, IA-52241 To whom correspondence should be addressed: Email: [email protected] (M.J.W.); yi- [email protected] (Y.X.); Email: [email protected] (P.B.M.) This PDF file includes: Materials and Methods References Fig. S1. miR-138 regulates SIN3A in a dose-dependent and site-specific manner. Fig. S2. miR-138 regulates endogenous SIN3A protein expression. Fig. S3. miR-138 regulates endogenous CFTR protein expression in Calu-3 cells. Fig. S4. miR-138 regulates endogenous CFTR protein expression in primary human airway epithelia. Fig. S5. miR-138 regulates CFTR expression in HeLa cells. Fig. S6. miR-138 regulates CFTR expression in HEK293T cells. Fig. S7. HeLa cells exhibit CFTR channel activity. Fig. S8. miR-138 improves CFTR processing. Fig. S9. miR-138 improves CFTR-ΔF508 processing. Fig. S10. SIN3A inhibition yields partial rescue of Cl- transport in CF epithelia. -
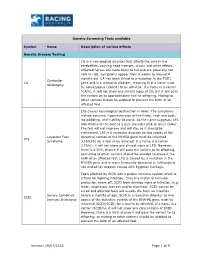
19/07/2018 Page 1 of 9 Genetic Screening Tests Available Symbol
Genetic Screening Tests available Symbol Name Description of variant effects Genetic Disease Testing CA is a neurological disorder that affects the cells in the cerebellum, causing head tremors, ataxia and other effects. Affected horses are more likely to fall and are generally not safe to ride. Symptoms appear from 6 weeks to around 4 months old. CA has been linked to a mutation in the TOE1 Cerebellar CA gene and is a recessive disorder, meaning that a horse must Abiotrophy be homozygous (CA/CA) to be affected. If a horse is a carrier (CA/n), it will not show any clinical signs of CA, but it will pass the variant on to approximately half its offspring. Mating to other carriers should be avoided to prevent the birth of an affected foal. LFS causes neurological dysfunction in foals. The symptoms include seizures, hyperextension of the limbs, neck and back, leg paddling, and inability to stand. As the name suggests, LFS also dilutes to the coat to a pale lavender pink or silver colour. The foal will not improve and will die, so it should be euthanised. LFS is a recessive disorder so two copies of the Lavender Foal defective version of the MYO5A gene must be inherited LFS Syndrome (LFS/LFS) for a foal to be affected. If a horse is a carrier (LFS/n), it will not show any clinical signs of LFS. However, there is a 50% chance it will pass the variant to its offspring, so mating to other carriers should be avoided to prevent the birth of an affected foal. -

Noelia Díaz Blanco
Effects of environmental factors on the gonadal transcriptome of European sea bass (Dicentrarchus labrax), juvenile growth and sex ratios Noelia Díaz Blanco Ph.D. thesis 2014 Submitted in partial fulfillment of the requirements for the Ph.D. degree from the Universitat Pompeu Fabra (UPF). This work has been carried out at the Group of Biology of Reproduction (GBR), at the Department of Renewable Marine Resources of the Institute of Marine Sciences (ICM-CSIC). Thesis supervisor: Dr. Francesc Piferrer Professor d’Investigació Institut de Ciències del Mar (ICM-CSIC) i ii A mis padres A Xavi iii iv Acknowledgements This thesis has been made possible by the support of many people who in one way or another, many times unknowingly, gave me the strength to overcome this "long and winding road". First of all, I would like to thank my supervisor, Dr. Francesc Piferrer, for his patience, guidance and wise advice throughout all this Ph.D. experience. But above all, for the trust he placed on me almost seven years ago when he offered me the opportunity to be part of his team. Thanks also for teaching me how to question always everything, for sharing with me your enthusiasm for science and for giving me the opportunity of learning from you by participating in many projects, collaborations and scientific meetings. I am also thankful to my colleagues (former and present Group of Biology of Reproduction members) for your support and encouragement throughout this journey. To the “exGBRs”, thanks for helping me with my first steps into this world. Working as an undergrad with you Dr. -

Horse Coat Color Test Results Dt34839
HORSE COAT COLOR TEST RESULTS TONI PERDEW Case: DT34839 3005 LEXINGTON CT Date Received: 05-Aug-2013 BEDFORD, IA 50833 Report Date: 07-Aug-2013 Report ID: 3263-6964-9709-4059 Verify report at https://www.vgl.ucdavis.edu/myvgl/verify.html Horse: HI JACD SILVER Reg: 5220615 DOB: 05/31/2009 Breed: QH Sex:M Alt. ID: Sire: MAINLY MERLIN Reg: Dam: BAILEYS BADLAND BUCK Reg: W10 DOMINANT RED FACTOR Not requested. Not requested. WHITE SPLASHED AGOUTI Not requested. Not requested. WHITE CREAM Not requested. TOBIANO Not requested. PEARL Not requested. LEOPARD Not requested. SILVER Not requested. GRAY Not requested. Horse is homozygous for the Dun gene. All DUN ROAN Not requested. D/D offspring should be dun dilute. CHAMPAGNE Not requested. LETHAL WHITE Not requested. OVERO SABINO 1 Not requested. For more details on horse coat color tests, please visit: www.vgl.ucdavis.edu/services/coatcolorhorse.php Tests for Gray, Leopard/Appalossa and Lethal White Overo are performed under license. Horse Coat Color Results with Explanations Red Factor Silver e/e - Only the red factor detected. Basic color is sorrel or chestnut in the absence of other N/N - No evidence of the altered sequence detected. modifying genes. N/Z - One copy of the altered sequence detected. Black-based horses will be chocolate with flaxen E/e - Both black and red factors detected. Either E or e transmitted to offspring. Basic color is or lightened mane and tail. Bay-based horses will have lightened black pigment on lower legs, mane black, bay or brown in the absence of other modifying genes. -

Human Lectins, Their Carbohydrate Affinities and Where to Find Them
biomolecules Review Human Lectins, Their Carbohydrate Affinities and Where to Review HumanFind Them Lectins, Their Carbohydrate Affinities and Where to FindCláudia ThemD. Raposo 1,*, André B. Canelas 2 and M. Teresa Barros 1 1, 2 1 Cláudia D. Raposo * , Andr1 é LAQVB. Canelas‐Requimte,and Department M. Teresa of Chemistry, Barros NOVA School of Science and Technology, Universidade NOVA de Lisboa, 2829‐516 Caparica, Portugal; [email protected] 12 GlanbiaLAQV-Requimte,‐AgriChemWhey, Department Lisheen of Chemistry, Mine, Killoran, NOVA Moyne, School E41 of ScienceR622 Co. and Tipperary, Technology, Ireland; canelas‐ [email protected] NOVA de Lisboa, 2829-516 Caparica, Portugal; [email protected] 2* Correspondence:Glanbia-AgriChemWhey, [email protected]; Lisheen Mine, Tel.: Killoran, +351‐212948550 Moyne, E41 R622 Tipperary, Ireland; [email protected] * Correspondence: [email protected]; Tel.: +351-212948550 Abstract: Lectins are a class of proteins responsible for several biological roles such as cell‐cell in‐ Abstract:teractions,Lectins signaling are pathways, a class of and proteins several responsible innate immune for several responses biological against roles pathogens. such as Since cell-cell lec‐ interactions,tins are able signalingto bind to pathways, carbohydrates, and several they can innate be a immuneviable target responses for targeted against drug pathogens. delivery Since sys‐ lectinstems. In are fact, able several to bind lectins to carbohydrates, were approved they by canFood be and a viable Drug targetAdministration for targeted for drugthat purpose. delivery systems.Information In fact, about several specific lectins carbohydrate were approved recognition by Food by andlectin Drug receptors Administration was gathered for that herein, purpose. plus Informationthe specific organs about specific where those carbohydrate lectins can recognition be found by within lectin the receptors human was body.