Online Resources Isolation and Characterization of Species-Specific
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Population, Distribution and Conservation Status of Sitatunga (Tragelaphus Spekei) (Sclater) in Selected Wetlands in Uganda
POPULATION, DISTRIBUTION AND CONSERVATION STATUS OF SITATUNGA (TRAGELAPHUS SPEKEI) (SCLATER) IN SELECTED WETLANDS IN UGANDA Biological -Life history Biological -Ecologicl… Protection -Regulation of… 5 Biological -Dispersal Protection -Effectiveness… 4 Biological -Human tolerance Protection -proportion… 3 Status -National Distribtuion Incentive - habitat… 2 Status -National Abundance Incentive - species… 1 Status -National… Incentive - Effect of harvest 0 Status -National… Monitoring - confidence in… Status -National Major… Monitoring - methods used… Harvest Management -… Control -Confidence in… Harvest Management -… Control - Open access… Harvest Management -… Control of Harvest-in… Harvest Management -Aim… Control of Harvest-in… Harvest Management -… Control of Harvest-in… Tragelaphus spekii (sitatunga) NonSubmitted Detrimental to Findings (NDF) Research and Monitoring Unit Uganda Wildlife Authority (UWA) Plot 7 Kira Road Kamwokya, P.O. Box 3530 Kampala Uganda Email/Web - [email protected]/ www.ugandawildlife.org Prepared By Dr. Edward Andama (PhD) Lead consultant Busitema University, P. O. Box 236, Tororo Uganda Telephone: 0772464279 or 0704281806 E-mail: [email protected] [email protected], [email protected] Final Report i January 2019 Contents ACRONYMS, ABBREVIATIONS, AND GLOSSARY .......................................................... vii EXECUTIVE SUMMARY ....................................................................................................... viii 1.1Background ........................................................................................................................... -

A Scoping Review of Viral Diseases in African Ungulates
veterinary sciences Review A Scoping Review of Viral Diseases in African Ungulates Hendrik Swanepoel 1,2, Jan Crafford 1 and Melvyn Quan 1,* 1 Vectors and Vector-Borne Diseases Research Programme, Department of Veterinary Tropical Disease, Faculty of Veterinary Science, University of Pretoria, Pretoria 0110, South Africa; [email protected] (H.S.); [email protected] (J.C.) 2 Department of Biomedical Sciences, Institute of Tropical Medicine, 2000 Antwerp, Belgium * Correspondence: [email protected]; Tel.: +27-12-529-8142 Abstract: (1) Background: Viral diseases are important as they can cause significant clinical disease in both wild and domestic animals, as well as in humans. They also make up a large proportion of emerging infectious diseases. (2) Methods: A scoping review of peer-reviewed publications was performed and based on the guidelines set out in the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) extension for scoping reviews. (3) Results: The final set of publications consisted of 145 publications. Thirty-two viruses were identified in the publications and 50 African ungulates were reported/diagnosed with viral infections. Eighteen countries had viruses diagnosed in wild ungulates reported in the literature. (4) Conclusions: A comprehensive review identified several areas where little information was available and recommendations were made. It is recommended that governments and research institutions offer more funding to investigate and report viral diseases of greater clinical and zoonotic significance. A further recommendation is for appropriate One Health approaches to be adopted for investigating, controlling, managing and preventing diseases. Diseases which may threaten the conservation of certain wildlife species also require focused attention. -

Mammal Species Richness at a Catena and Nearby Waterholes During a Drought, Kruger National Park, South Africa
diversity Article Mammal Species Richness at a Catena and Nearby Waterholes during a Drought, Kruger National Park, South Africa Beanélri B. Janecke Animal, Wildlife & Grassland Sciences, University of the Free State, 205 Nelson Mandela Road, Park West, Bloemfontein 9301, South Africa; [email protected]; Tel.: +27-51-401-9030 Abstract: Catenas are undulating hillslopes on a granite geology characterised by different soil types that create an environmental gradient from crest to bottom. The main aim was to determine mammal species (>mongoose) present on one catenal slope and its waterholes and group them by feeding guild and body size. Species richness was highest at waterholes (21 species), followed by midslope (19) and sodic patch (16) on the catena. Small differences observed in species presence between zones and waterholes and between survey periods were not significant (p = 0.5267 and p = 0.9139). In total, 33 species were observed with camera traps: 18 herbivore species, 10 carnivores, two insectivores and three omnivores. Eight small mammal species, two dwarf antelopes, 11 medium, six large and six mega-sized mammals were observed. Some species might not have been recorded because of drought, seasonal movement or because they travelled outside the view of cameras. Mammal presence is determined by food availability and accessibility, space, competition, distance to water, habitat preferences, predators, body size, social behaviour, bound to territories, etc. The variety in body size and feeding guilds possibly indicates a functioning catenal ecosystem. This knowledge can be beneficial in monitoring and conservation of species in the park. Keywords: catena ecosystem; ephemeral mud wallows; habitat use; mammal variety; Skukuza area; Citation: Janecke, B.B. -

Mixed-Species Exhibits with Pigs (Suidae)
Mixed-species exhibits with Pigs (Suidae) Written by KRISZTIÁN SVÁBIK Team Leader, Toni’s Zoo, Rothenburg, Luzern, Switzerland Email: [email protected] 9th May 2021 Cover photo © Krisztián Svábik Mixed-species exhibits with Pigs (Suidae) 1 CONTENTS INTRODUCTION ........................................................................................................... 3 Use of space and enclosure furnishings ................................................................... 3 Feeding ..................................................................................................................... 3 Breeding ................................................................................................................... 4 Choice of species and individuals ............................................................................ 4 List of mixed-species exhibits involving Suids ........................................................ 5 LIST OF SPECIES COMBINATIONS – SUIDAE .......................................................... 6 Sulawesi Babirusa, Babyrousa celebensis ...............................................................7 Common Warthog, Phacochoerus africanus ......................................................... 8 Giant Forest Hog, Hylochoerus meinertzhageni ..................................................10 Bushpig, Potamochoerus larvatus ........................................................................ 11 Red River Hog, Potamochoerus porcus ............................................................... -

List of 28 Orders, 129 Families, 598 Genera and 1121 Species in Mammal Images Library 31 December 2013
What the American Society of Mammalogists has in the images library LIST OF 28 ORDERS, 129 FAMILIES, 598 GENERA AND 1121 SPECIES IN MAMMAL IMAGES LIBRARY 31 DECEMBER 2013 AFROSORICIDA (5 genera, 5 species) – golden moles and tenrecs CHRYSOCHLORIDAE - golden moles Chrysospalax villosus - Rough-haired Golden Mole TENRECIDAE - tenrecs 1. Echinops telfairi - Lesser Hedgehog Tenrec 2. Hemicentetes semispinosus – Lowland Streaked Tenrec 3. Microgale dobsoni - Dobson’s Shrew Tenrec 4. Tenrec ecaudatus – Tailless Tenrec ARTIODACTYLA (83 genera, 142 species) – paraxonic (mostly even-toed) ungulates ANTILOCAPRIDAE - pronghorns Antilocapra americana - Pronghorn BOVIDAE (46 genera) - cattle, sheep, goats, and antelopes 1. Addax nasomaculatus - Addax 2. Aepyceros melampus - Impala 3. Alcelaphus buselaphus - Hartebeest 4. Alcelaphus caama – Red Hartebeest 5. Ammotragus lervia - Barbary Sheep 6. Antidorcas marsupialis - Springbok 7. Antilope cervicapra – Blackbuck 8. Beatragus hunter – Hunter’s Hartebeest 9. Bison bison - American Bison 10. Bison bonasus - European Bison 11. Bos frontalis - Gaur 12. Bos javanicus - Banteng 13. Bos taurus -Auroch 14. Boselaphus tragocamelus - Nilgai 15. Bubalus bubalis - Water Buffalo 16. Bubalus depressicornis - Anoa 17. Bubalus quarlesi - Mountain Anoa 18. Budorcas taxicolor - Takin 19. Capra caucasica - Tur 20. Capra falconeri - Markhor 21. Capra hircus - Goat 22. Capra nubiana – Nubian Ibex 23. Capra pyrenaica – Spanish Ibex 24. Capricornis crispus – Japanese Serow 25. Cephalophus jentinki - Jentink's Duiker 26. Cephalophus natalensis – Red Duiker 1 What the American Society of Mammalogists has in the images library 27. Cephalophus niger – Black Duiker 28. Cephalophus rufilatus – Red-flanked Duiker 29. Cephalophus silvicultor - Yellow-backed Duiker 30. Cephalophus zebra - Zebra Duiker 31. Connochaetes gnou - Black Wildebeest 32. Connochaetes taurinus - Blue Wildebeest 33. Damaliscus korrigum – Topi 34. -
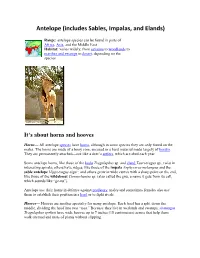
Antelope (Includes Sables, Impalas, and Elands)
Antelope (includes Sables, Impalas, and Elands) Range: antelope species can be found in parts of Africa, Asia, and the Middle East Habitat: varies widely, from savanna to woodlands to marshes and swamps to desert, depending on the species It’s about horns and hooves Horns— All antelope species have horns, although in some species they are only found on the males. The horns are made of a bony core, encased in a hard material made largely of keratin. They are permanently attached—not like a deer’s antlers, which are shed each year. Some antelope horns, like those of the kudu Tragelaphus sp. and eland Taurotragus sp., twist in interesting spirals; others have ridges, like those of the impala Aephyceros melampus and the sable antelope Hippotragus niger; and others grow in wide curves with a sharp point on the end, like those of the wildebeest Connochaetes sp. (also called the gnu, a name it gets from its call, which sounds like “ge-nu”). Antelope use their horns in defense against predators; males and sometimes females also use them to establish their position in a herd or to fight rivals Hooves— Hooves are another specialty for many antelope. Each hoof has a split down the middle, dividing the hoof into two “toes.” Because they live in wetlands and swamps, sitatungas Tragelaphus spekeii have wide hooves up to 7 inches (18 centimeters) across that help them walk on mud and mats of plants without slipping. Nile lechwes Kobus magaceros, which also live in swampy areas, have long, pointed hooves to give them sure footing in the water. -

Species-Packing of Grazers in the Mole National Park, Ghana
Species-Packing of Grazers in the Mole National Park, Ghana Dakwa, K. B. Department of Conservation Biology and Entomology School of Biological Sciences, University of Cape Coast, Ghana *Corresponding Author: [email protected] / [email protected] Abstract Hutchinson’s body weight ratio (WR) theory was used to determine the degree of species-packing of 22 species of grazers in Mole National Park (MNP). The hypotheses, which suggest that facilitation is more likely to occur at a WR greater than 2.0, competition at WR less than 2.0, and coexistence at WR equals 2.0 were tested by regressing the natural logarithm of the body mass of a grazer against its rank number in the grazer assemblage. The results indicated competition in the grazer assemblage at MNP as its WR is 1.40 and therefore grazers are tightly packed. However, as several species with similar body weight coexisted at MNP, the Hutchinson’s Rule could not be supported. Habitat heterogeneity rather than the size of conservation area related to species-packing and MNP with its low habitat heterogeneity showed a low degree of species-packing. Possible explanations have been advanced for existing ecological holes within the assemblage. Species-packing could still be a reasonable measure of the characteristics, dynamics, interactions and the patterns of assemble in animal communities. It could also be used to predict the effect of animal species loss or arrival on the stability of a natural ecosystem and provide useful guidelines in planning herbivore conservation measures in protected areas. Introduction in the assemblages of sympatric species in an area. -

Tuberculosis in Kafue Lechwe (Kobus Leche Kafuensis) and in a Bushbuck (Tragelaphus Scriptus) on a Game Ranch in Central Province, Zambia
Case report — Gevalverslag Tuberculosis in Kafue lechwe (Kobus leche kafuensis) and in a bushbuck (Tragelaphus scriptus) on a game ranch in Central Province, Zambia U Ziegera*, G S Pandeyb, N P J Kriekc and A E Cauldwella bacteriosis, 2 in Kafue lechwe and 1 in a ABSTRACT bushbuck from a private game ranch near Mycobacteriosis was diagnosed for the first time outside a national park in free-ranging Lusaka. The pathological and histological wild animals on a game ranch in Zambia. A Kafue lechwe (Kobus leche kafuensis) was found lesions were typical of tuberculosis in all 3 dead with tuberculous lesions on a ranch near Lusaka. Acid-fast bacilli were found in the cases; however, mycobacteria could only affected organs. Mycobacteria were isolated from these tissues. A bushbuck (Tragelaphus be isolated from 1 of the Kafue lechwe. scriptus) was found dead on the same ranch with multiple superficial abscesses in the neck Unfortunately, appropriate biochemical region, extensive granulomatous lesions in the lung, the bronchial and mediastinal lymph tests were not available at the time to nodes and several nodular lesions in the spleen. Few acid-fast bacilli were found in the exudate from the abscesses and lesions in the affected organs. Histologically the lesions confirm without doubt that the isolate resembled those of tuberculosis, but mycobacteria could not be isolated. In addition, 1 Kafue was M bovis. The strong suspicion, lechwe among 37 wild ungulates of 13 species shot on the ranch showed typical tuberculous however, that bovine tuberculosis is lesions in the lungs, but the diagnosis was not confirmed by bacterial isolation. -

Title 50 Part 17
U Title 50—Wildlife and Fisheries Ihe information of the reader. In the annual all other appropriate rules in Parts 17. 217 revision and compilation of this title, the through 227. and 402 still apply to that PART 17—ENDANGERED AND following information may be amended species. In addition, there may be other rules THREATENED WILDLIFE AND PLANTS without public notice: the spelling of species' in this title that relate to such wildlife, e.g.. names, historical range, footnotes, references pon-of-entry requirements. It is not intended to certain other applicable portions of this that the references m the "Special rules" title, synonyms, and more current names. In column list all the regulations of the two Subpart B—Lists any of these revised entries, neither the Services which might apply to the species or § 17.11 Endangered and threatened species, as defined in paragraph (b) of this to the regulations of other Federal agencies wildlife. section, nor its status may be changed without or Stale or local governments. (a) The list in this section contains the following the procedures of Part 424 of this (g) The listing of a particular taxon names of all species of wildlife which have title. includes all lower taxonomic units. For been determined by Ihe Services to be (e) The "historic range" indicates the example, the genus H\lobaies (gibbons) is Endangered or Threatened. It also contains known general distribution of the species or listed as Endangered throughout its entire the names of species of wildlife treated as subspecies as reported in the current scientific range (China. -

Suiform Soundings
Suiform Soundings Newsletter of the WPSG, ISSN: 1446-991X PSG and HSG Volume 11(2) August 2012 Suiform Soundings is the newsletter of the IUCN/SSC Wild Pig, Peccary, and Hippo Specialist Groups. This newsletter is electronically available at: http://data.iucn.org/themes/ssc/sgs/pphsg/Suiform%20soundings/Newsletter.htm Photo front page: An impressive male warthog, Phacochoerus africanus, in the Bale Mountains National Park, southern Ethiopia. Photo courtesy of Jeffrey Graham TABLE OF CONTENTS EDITORIAL by Anne-Marie Stewart 3 A “Chacoan Peccary Workshop” to re-evaluate the species’ Red List Status and develop a Conservation Ac- tion Plan by Harald Beck and Mariana Altrichter 3 A brief summary of the Chacoan Peccary in Paraguay, and research and conservation needs for the species. by Juan Campos 4 News From the SSC Peccary Specialist Group 6 PAPERS AND COMMUNICATIONS Ovarian functional anatomy in the Wild Boar (Sus scrofa) from North-eastern Spain by Pedro Mayor, Gregorio Mentaberre, Ignasi Marco, Santiago Lavín, Francesc Closa, Víctor Sazatornil, Carlos López-Plana and Manel López-Béjar 7 IUCN red listing status for peccaries in Brazil: Should the global status of the white-lipped peccary be up- graded? by A Keuroghlian, ALJ Desbiez, BM Beisiegel, A Gatti, AR Mendes Pontes, CB Campos, CF Tófoli, EA Moraes Jr, GM Pinho, LP Cordeiro, TS Santos Jr, AA Morais, PR Mangini, K Flesher, LF Rodrigues and LB Almeida 14 Mexico has raised the legal status of the white-lipped peccary by Rafael Reyna-Hurtado, Rodrigo A. Medellín and Eduardo J. Naranjo 16 An eyeless Eurasian Wild Pig (Sus scrofa) surviving in northern Laos by Jan F. -

Antelopes, Gazelles, Cattle, Goats, Sheep, and Relatives
© Copyright, Princeton University Press. No part of this book may be distributed, posted, or reproduced in any form by digital or mechanical means without prior written permission of the publisher. INTRODUCTION RECOGNITION The family Bovidae, which includes Antelopes, Cattle, Duikers, Gazelles, Goats, and Sheep, is the largest family within Artiodactyla and the most diverse family of ungulates, with more than 270 recent species. Their common characteristic is their unbranched, non-deciduous horns. Bovids are primarily Old World in their distribution, although a few species are found in North America. The name antelope is often used to describe many members of this family, but it is not a definable, taxonomically based term. Shape, size, and color: Bovids encompass an extremely wide size range, from the minuscule Royal Antelope and the Dik-diks, weighing as little as 2 kg and standing 25 to 35 cm at the shoulder, to the Asian Wild Water Buffalo, which weighs as much as 1,200 kg, and the Gaur, which measures up to 220 cm at the shoulder. Body shape varies from relatively small, slender-limbed, and thin-necked species such as the Gazelles to the massive, stocky wild cattle (fig. 1). The forequarters may be larger than the hind, or the reverse, as in smaller species inhabiting dense tropical forests (e.g., Duikers). There is also a great variety in body coloration, although most species are some shade of brown. It can consist of a solid shade, or a patterned pelage. Antelopes that rely on concealment to avoid predators are cryptically colored. The stripes and blotches seen on the hides of Bushbuck, Bongo, and Kudu also function as camouflage by helping to disrupt the animals’ outline. -

The Case of White Eared Kob in Ethiopia and South Sudan Landscape
Conservation of transboundary migratory mammals: the case of white eared kob in Ethiopia and South Sudan landscape Principal investigator: Cherie Enawgaw Beyene Abstract Migratory species in particular mammals are in sharp decline across the globe. In 2007, world second largest migratory animals consisting of larger proportion of white eared kob (Kobus kob leucotis) with other many large mammals were reported crossing the border of Ethiopia from Southern Sudan surviving the two decade of civil war in the region. While mass migration of mammals in the Boma-Jonglei Ecosystem is known from 1970 and 1980 studies, it was believed these migrations were thought to be decimated by civil war. Little is known how such large population of wildlife survive; it is likely that the area was dominated by Sudan People's Liberation Army (SPLA) little affected by large public. While this is good news, changes made in Social structure and human development following the independence of south Sudan may threat such spectacular migration. In both countries, foreign investment for large scale agriculture and Construction of Dam for hydroelectric power plant is increasing. Without considering critical habitat and migration corridors, such development may case a quick collapse due to uncontrolled human impact. In March 2013, supported by Conservation of Migratory Species (CMS) we collared 18 white eared kob providing a very useful information on the movement pattern and corridors of the white eared kob over 230km air distance landscape between Ethiopia and south Sudan (85,000 km2 area as combined home range). So far the GPS coordinates (fixes every 4 hrs) is downloaded from web site from the programmed satellite collars.