Control of Relb During Dendritic Cell Activation
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Genomic Analyses of PMBL Reveal New Drivers and Mechanisms of Sensitivity to PD-1 Blockade
Regular Article LYMPHOID NEOPLASIA Genomic analyses of PMBL reveal new drivers and mechanisms of sensitivity to PD-1 blockade Bjoern Chapuy,1,2,* Chip Stewart,3,* Andrew J. Dunford,3,* Jaegil Kim,3 Kirsty Wienand,1 Atanas Kamburov,3 Gabriel K. Griffin,4 Pei-Hsuan Chen,4 Ana Lako,4 Robert A. Redd,5 Claire M. Cote,1 Matthew D. Ducar,6 Aaron R. Thorner,6 Scott J. Rodig,4 Gad Getz,3,7,8,† Downloaded from https://ashpublications.org/blood/article-pdf/134/26/2369/1549702/bloodbld2019002067.pdf by Margaret Shipp on 30 December 2019 and Margaret A. Shipp1,† 1Department of Medical Oncology, Dana-Farber Cancer Institute, Harvard Medical School, Boston, MA; 2Department of Hematology and Oncology, University Medical Center Gottingen,¨ Gottingen,¨ Germany; 3Broad Institute of Harvard and Massachusetts Institute of Technology, Cambridge, MA; 4Department of Pathology, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA; 5Department of Data Sciences and 6Center for Cancer Genome Discovery, Dana-Farber Cancer Institute, Boston, MA; and 7Department of Pathology and 8Center for Cancer Research, Massachusetts General Hospital, Harvard Medical School, Boston, MA KEY POINTS Primary mediastinal large B-cell lymphomas (PMBLs) are aggressive tumors that typically present as large mediastinal masses in young women. PMBLs share clinical, transcriptional, l Comprehensive genomic analyses of and molecular features with classical Hodgkin lymphoma (cHL), including constitutive PMBL reveal new activation of nuclear factor kB (NF-kB), JAK/STAT signaling, and programmed cell death genetic drivers such protein 1 (PD-1)–mediated immune evasion. The demonstrated efficacy of PD-1 blockade in as ZNF217. relapsed/refractory PMBLs led to recent approval by the US Food and Drug Administration l High mutational and underscored the importance of characterizing targetable genetic vulnerabilities in this burden, MSI, and disease. -

Transcriptome Analysis of Human Diabetic Kidney Disease
ORIGINAL ARTICLE Transcriptome Analysis of Human Diabetic Kidney Disease Karolina I. Woroniecka,1 Ae Seo Deok Park,1 Davoud Mohtat,2 David B. Thomas,3 James M. Pullman,4 and Katalin Susztak1,5 OBJECTIVE—Diabetic kidney disease (DKD) is the single cases, mild and then moderate mesangial expansion can be leading cause of kidney failure in the U.S., for which a cure has observed. In general, diabetic kidney disease (DKD) is not yet been found. The aim of our study was to provide an considered a nonimmune-mediated degenerative disease unbiased catalog of gene-expression changes in human diabetic of the glomerulus; however, it has long been noted that kidney biopsy samples. complement and immunoglobulins sometimes can be de- — tected in diseased glomeruli, although their role and sig- RESEARCH DESIGN AND METHODS Affymetrix expression fi arrays were used to identify differentially regulated transcripts in ni cance is not clear (4). 44 microdissected human kidney samples. The DKD samples were The understanding of DKD has been challenged by multi- significant for their racial diversity and decreased glomerular ple issues. First, the diagnosis of DKD usually is made using filtration rate (~20–30 mL/min). Stringent statistical analysis, using clinical criteria, and kidney biopsy often is not performed. the Benjamini-Hochberg corrected two-tailed t test, was used to According to current clinical practice, the development of identify differentially expressed transcripts in control and diseased albuminuria in patients with diabetes is sufficient to make the glomeruli and tubuli. Two different Web-based algorithms were fi diagnosis of DKD (5). We do not understand the correlation used to de ne differentially regulated pathways. -
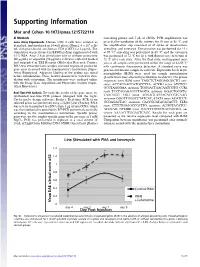
Supporting Information
Supporting Information Mor and Cohen 10.1073/pnas.1215722110 SI Methods containing primer and 5 μL of cDNA. PCR amplification was Gene Array Experiments. Human CD4 T cells were isolated as preceded by incubation of the mixture for 10 min at 95 °C, and described, and incubated in 24-well plates (Nunc), 4 × 106 cells/ the amplification step consisted of 45 cycles of denaturation, mL with plate-bound anti-human CD3 (OKT3) at 2 μg/mL. The annealing, and extension. Denaturation was performed for 15 s stimulation was performed in RPMI medium supplemented with at 95 °C; annealing was performed in 60 °C; and the extension 0.1% BSA. After 2 h of stimulation with or without cefuroxime was performed at 72 °C for 20 s, with fluorescence detection at (50 μg/mL) or ampicillin (50 μg/mL), cells were collected washed 72 °C after each cycle. After the final cycle, melting-point anal- and suspended in TRI Reagent (Molecular Research Center). yses of all samples were performed within the range of 62–95 °C RNA was extracted from samples and used to prepare probes for with continuous fluorescence detection. A standard curve was gene array in accord with the manufacturer’s instructions (Super- generated from one sample in each run. Expression levels of β2- Array Bioscience). Adequate labeling of the probes was tested microglobulin (B2M) were used for sample normalization before hybridization. Three healthy donors were tested in stim- (β-actin levels were affected by cefuroxime treatment). The primer ulation with cefuroxime. The membranes were analyzed online sequences were B2M sense TAGCTCTAGGAGGGCTG anti- with the Image Data Acquisition and Expression Analysis (Super- sense ACCACAACCATGCCTTA; ACVR2 sense ATCTCC- Array Bioscience). -

Genome-Wide Identification of Genes Regulating DNA Methylation Using Genetic Anchors for Causal Inference
bioRxiv preprint doi: https://doi.org/10.1101/823807; this version posted October 30, 2019. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY 4.0 International license. Genome-wide identification of genes regulating DNA methylation using genetic anchors for causal inference Paul J. Hop1,2, René Luijk1, Lucia Daxinger3, Maarten van Iterson1, Koen F. Dekkers1, Rick Jansen4, BIOS Consortium5, Joyce B.J. van Meurs6, Peter A.C. ’t Hoen 7, M. Arfan Ikram8, Marleen M.J. van Greevenbroek9,10, Dorret I. Boomsma11, P. Eline Slagboom1, Jan H. Veldink2, Erik W. van Zwet12, Bastiaan T. Heijmans1* 1 Molecular Epidemiology, Department of Biomedical Data Sciences, Leiden University Medical Center, Leiden, 2333 ZC, The Netherlands 2 Department of Neurology, UMC Utrecht Brain Center, University Medical Centre Utrecht, Utrecht University, Utrecht 3584 CG, Netherlands 3 Department of Human Genetics, Leiden University Medical Center, Leiden, 2333 ZC, The Netherlands 4 Department of Psychiatry, Amsterdam UMC, Vrije Universiteit Amsterdam, Amsterdam Neuroscience, Amsterdam, 1081 HV, The Netherlands 5 Biobank-based Integrated Omics Study Consortium. For a complete list of authors, see the acknowledgements. 6 Department of Internal Medicine, Erasmus Medical Centre, Rotterdam, The Netherlands. 7 Centre for Molecular and Biomolecular Informatics, Radboud Institute for Molecular Life Sciences, Radboud University -

The Role of the NFAT Signalling Pathway in Diffuse Large B-Cell Lymphoma
The role of the NFAT signalling pathway in Diffuse Large B-cell Lymphoma Holly White Doctor of Philosophy Thesis September 2015 Supervisors: Dr Chris Bacon, Professor Neil Perkins & Dr Vikki Rand Northern Institute for Cancer Research Word count: 66,024 1 Abstract Diffuse Large B-Cell Lymphomas (DLBCL) are common, aggressive malignancies of mature B-lymphocytes that represent ~40% of lymphomas. Despite the widespread use of combined immunochemotherapy, approximately 50% of patients with DLBCL die from their disease. The two main DLBCL subgroups resemble activated B cells (ABC) or germinal centre B cells (GCB), where patients with ABC-DLBCL have significantly worse outcome. There is urgent need for novel therapeutic strategies in the treatment of DLBCL, which requires a better understanding of the molecular pathways upon which tumours depend. Accumulating evidence suggests that the signalling networks promoting and sustaining DLBCL derive from dysregulation of the normal pathways controlling B-lymphocyte activation and differentiation. There is increasing evidence indicating important roles for the NFAT family of transcription factors in DLBCL. Constitutively-active nuclear NFAT2 has been demonstrated in approximately 40% of primary DLBCL samples and NFAT has been shown to regulate a small number of genes associated with DLBCL growth/survival. This project investigated the role of NFAT in DLBCL. Nuclear localisation and activation of NFAT family members were characterised in a panel of DLBCL cell lines and chemical inhibition of calcineurin/NFAT, using Cyclosporin A (CsA), indicated dependency on the calcineurin/NFAT pathway for survival. Gene expression microarray analysis performed in DLBCL cell lines treated with CsA revealed potential NFAT target genes involved in the tumour microenvironment and anergy. -

Recurrent Genetic Mutations in Lymphoid Malignancies
Digital Comprehensive Summaries of Uppsala Dissertations from the Faculty of Medicine 1312 Recurrent Genetic Mutations in Lymphoid Malignancies EMMA YOUNG ACTA UNIVERSITATIS UPSALIENSIS ISSN 1651-6206 ISBN 978-91-554-9850-4 UPPSALA urn:nbn:se:uu:diva-314956 2017 Dissertation presented at Uppsala University to be publicly examined in Rudbecksalen, Dag Hammarskjölds Väg 20, Uppsala, Friday, 5 May 2017 at 13:15 for the degree of Doctor of Philosophy (Faculty of Medicine). The examination will be conducted in English. Faculty examiner: Professor Dimitar Efremov (Molecular Hematology Group of the International Centre for Genetic Engineering and Biotechnology (ICGEB) in Trieste, Italy). Abstract Young, E. 2017. Recurrent Genetic Mutations in Lymphoid Malignancies. Digital Comprehensive Summaries of Uppsala Dissertations from the Faculty of Medicine 1312. 82 pp. Uppsala: Acta Universitatis Upsaliensis. ISBN 978-91-554-9850-4. In recent years, the genetic landscape of B-cell derived lymphoid malignancies, including chronic lymphocytic leukemia (CLL), has been rapidly unraveled, identifying recurrent genetic mutations with potential clinical impact. Interestingly, ~30% of all CLL patients can be assigned to more homogeneous subsets based on the expression of a similar or “stereotyped” B-cell receptor (BcR). Considering that biased distribution of genetic mutations was recently indicated in specific stereotyped subsets, in paper I, we screened 565 subset cases, preferentially assigned to clinically aggressive subsets, and confirm the SF3B1 mutational bias in subset #2 (45%), but also report on similarly marked enrichment in subset #3 (46%). In contrast, NOTCH1 mutations were predominantly detected in subsets #1, #8, #59 and #99 (22-34%). This data further highlights a subset-biased acquisition of genetic mutations in the pathogenesis of at least certain subsets. -

Supplemental Materials
SUPPLEMENTARY DATA shRNA-containing lentiviral vectors. The shRNAs for each target genes were designed with the aid of web-based Invitrogen Block-It program. The oligonucleotides containing Fra-1, c-Jun or slug shRNA sequences were inserted into pLV-shRNA vector (Biosettia, San Diego, CA) according to manufacturer’s protocol. The Fra-1 mRNA target sequences are 5’- GGAGACTGACAAACTGGAAGA-3’ and 5’-GGATGGTACAGCCTCATTTCC-3’. The c-Jun mRNA target sequences are 5’-GGAACAGGTGGCACAGCTTAA-3’ and 5’- GGCACAGCTTAAACAGAAAGT-3’. The slug mRNA target sequences are 5’- GCATTTGCAGACAGGTCAAAT-3’ and 5’-GCTCATCTGCAGACCCATTCT-3’. Slug promoter construction. The slug promoter (GeneBank Accession number AB300659) was synthesized by PCR using genomic DNA isolated from MDA-MB-231 cells. The synthesized fragment was subcloned into pGL4-luciferase reporter gene plasmid (Promega, Madison, WI) and sequence confirmed by automated DNA sequencing. To generate AP1 mutant, the AP1 consensus sequence in the slug promoter was destroyed by introducing mutation at nucleotide 1009-1011 (GAC AGA). To determine the promoter activity, wild-type or mutant slug promoter reporter gene construct was transfected into MCF-7, MDA-MB-231, MDA-MB-436 or ZR-75-1 cells for 36 hrs and cell lysates collected to measure luciferase activity using Lipofectamine (Invitrogen, Carlsbad, CA). To determine the effect of Fra-1/c-Jun expression on slug promoter activity, the promoter reporter gene construct was co-transfected with expression vectors encoding Fra-1 and c-Jun into MCF-7 and ZR-75-1 cells for 48 hrs followed by the analysis of luciferase activity. Plasmid containing Renilla Luciferase gene was included during transfection to serve as an internal control and Dual Luciferase Assay System (Promega, Madison, WI) was used for measuring luciferase activity. -

Circulating Tumor DNA Reveals Genetics, Clonal Evolution, and Residual Disease in Classical Hodgkin Lymphoma
From www.bloodjournal.org by Fredrik Schjesvold on August 4, 2018. For personal use only. Plenary Paper LYMPHOID NEOPLASIA Circulating tumor DNA reveals genetics, clonal evolution, and residual disease in classical Hodgkin lymphoma Valeria Spina,1,* Alessio Bruscaggin,1,* Annarosa Cuccaro,2 Maurizio Martini,3 Martina Di Trani,4 Gabriela Forestieri,1 Martina Manzoni,5 Adalgisa Condoluci,1,6 Alberto Arribas,1 Lodovico Terzi-Di-Bergamo,1 Silvia Laura Locatelli,4 Elisa Cupelli,2 Luca Ceriani,6 Alden A. Moccia,6 Anastasios Stathis,6 Luca Nassi,7 Clara Deambrogi,7 Fary Diop,7 Francesca Guidetti,1 Alessandra Cocomazzi,3 Salvatore Annunziata,8 Vittoria Rufini,8 Alessandro Giordano,8 Antonino Neri,5,9 Renzo Boldorini,10 Bernhard Gerber,6 Francesco Bertoni,1,6 Michele Ghielmini,6 Georg Stussi,¨ 6 Armando Santoro,4,11 Franco Cavalli,1,6 Emanuele Zucca,6 Luigi Maria Larocca,3 Gianluca Gaidano,7 Stefan Hohaus,2,† Carmelo Carlo-Stella,4,11,† and Davide Rossi1,6,† 1Institute of Oncology Research, Bellinzona, Switzerland; 2Institute of Hematology and 3Division of Pathology, Policlinico Gemelli Foundation, Catholic University of the Sacred Heart, Rome, Italy; 4Humanitas Cancer Center, Humanitas Clinical and Research Center, Milan, Italy; 5Department of Oncology and Hemato-oncology, University of Milan, Milan, Italy; 6Oncology Institute of Southern Switzerland, Bellinzona, Switzerland; 7Division of Hematology, Department of Translational Medicine, University of Eastern Piedmont, Novara, Italy; 8Institute of Nuclear Medicine, Policlinico Gemelli Foundation, Catholic University of the Sacred Heart, Rome, Italy; 9Hematology Unit, Foundation Ca’ Granda IRCCS, Ospedale Maggiore Policlinico, Milan, Italy; 10Division of Pathology, Department of Health Science, University of Eastern Piedmont, Novara, Italy; and 11Department of Biomedical Sciences, Humanitas University, Milan, Italy KEY POINTS The rarity of neoplastic cells in the biopsy imposes major technical hurdles that have so far limited genomic studies in classical Hodgkin lymphoma (cHL). -

UNIVERSITY of CALIFORNIA Los Angeles Dissecting the Regulatory
UNIVERSITY OF CALIFORNIA Los Angeles Dissecting the Regulatory Strategies of NFkappaB RelA Target Genes in the Inflammatory Response A dissertation submitted in partial satisfaction of the requirements for the degree Doctor of Philosophy in Microbiology, Immunology, and Molecular Genetics by Kim Anh Ngo 2019 © Copyright by Kim Anh Ngo 2019 ABSTRACT OF THE DISSERTATION Dissecting the Regulatory Strategies of NFkappaB RelA Target Genes in the Inflammatory Response by Kim Anh Ngo Doctor of Philosophy in Microbiology, Immunology, and Molecular Genetics University of California, Los Angeles, 2019 Professor Alexander Hoffmann, Chair The NFkB family member RelA is a ubiquitously expressed potent transcriptional activator that is induced by exposure to pathogens and inflammatory cytokines to activate the expression of a large number of inflammatory and immune-response genes. Its nuclear activity is induced from a latent cytoplasmic pool by stimulus-responsive degradation of IkB proteins, and the complex signaling mechanisms that regulate its activity are well understood. Less well characterized are the mechanisms that allow nuclear NFkB RelA activity to select its target genes and produce gene-specific expression. While many genes have been identified to be potentially NFkB regulated, there is no database that lists the NFkB target genes in a particular physiological condition, defined cell types and stimulus. ii Chapter 1 presents a general overview of IKK-IkB-NFkB signaling system. Chapter 2 reports the primary study in this dissertation in which includes approaches such as biochemistry, molecular biology, mouse genetics, Next-Generation Sequencing, and mathematical modeling to dissect the regulatory strategies of NFkB RelA endogenous target genes in the inflammatory response. -

IKB Epsilon (NFKBIE) Rabbit Polyclonal Antibody Product Data
OriGene Technologies, Inc. 9620 Medical Center Drive, Ste 200 Rockville, MD 20850, US Phone: +1-888-267-4436 [email protected] EU: [email protected] CN: [email protected] Product datasheet for TA322729 IKB epsilon (NFKBIE) Rabbit Polyclonal Antibody Product data: Product Type: Primary Antibodies Applications: IHC, WB Recommended Dilution: ELISA: 1:1000-2000, WB: 1:200-1000,IHC: 1:25-100 Reactivity: Human, Mouse, Rat Host: Rabbit Isotype: IgG Clonality: Polyclonal Immunogen: Fusion protein corresponding to a region derived from 63-423 amino acids of human nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, epsilon Formulation: PBS pH7.3, 0.05% NaN3, 50% glycerol Concentration: lot specific Purification: Antigen affinity purification Conjugation: Unconjugated Storage: Store at -20°C as received. Stability: Stable for 12 months from date of receipt. Predicted Protein Size: 45 kDa Gene Name: NFKB inhibitor epsilon Database Link: NP_004547 Entrez Gene 18037 MouseEntrez Gene 316241 RatEntrez Gene 4794 Human O00221 This product is to be used for laboratory only. Not for diagnostic or therapeutic use. View online » ©2021 OriGene Technologies, Inc., 9620 Medical Center Drive, Ste 200, Rockville, MD 20850, US 1 / 2 IKB epsilon (NFKBIE) Rabbit Polyclonal Antibody – TA322729 Background: The protein encoded by this gene binds to components of NF-kappa-B; trapping the complex in the cytoplasm and preventing it from activating genes in the nucleus. Phosphorylation of the encoded protein targets it for destruction by the ubiquitin pathway; which activates NF- kappa-B by making it available to translocate to the nucleus. NFKBIE protein expression is up- regulated following NF-?B activation and during myelopoiesis. -

Assessment of Transmission Distortion on Chromosome 6P in Healthy Individuals Using Tagsnps
European Journal of Human Genetics (2009) 17, 1182 – 1189 & 2009 Macmillan Publishers Limited All rights reserved 1018-4813/09 $32.00 www.nature.com/ejhg ARTICLE Assessment of transmission distortion on chromosome 6p in healthy individuals using tagSNPs Pablo Sandro Carvalho Santos*,1,4, Johannes Ho¨hne2,4, Peter Schlattmann2, Inke R Ko¨nig3, Andreas Ziegler3, Barbara Uchanska-Ziegler1 and Andreas Ziegler1 1Institut fu¨r Immungenetik, Charite´-Universita¨tsmedizin Berlin, Berlin, Germany; 2Institut fu¨r Biometrie und klinische Epidemiologie, Charite´-Universita¨tsmedizin Berlin, Berlin, Germany; 3Institut fu¨r medizinische Biometrie und Statistik, Universita¨t zu Lu¨beck, Lu¨beck, Germany The best-documented example for transmission distortion (TD) to normal offspring are the t haplotypes on mouse chromosome 17. In healthy humans, TD has been described for whole chromosomes and for particular loci, but multiple comparisons have presented a statistical obstacle in wide-ranging analyses. Here we provide six high-resolution TD maps of the short arm of human chromosome 6 (Hsa6p), based on single-nucleotide polymorphism (SNP) data from 60 trio families belonging to two ethnicities that are available through the International HapMap Project. We tested all approximately 70 000 previously genotyped SNPs within Hsa6p by the transmission disequilibrium test. TagSNP selection followed by permutation testing was performed to adjust for multiple testing. A statistically significant evidence for TD was observed among male parents of European ancestry, due to strong and wide-ranging skewed segregation in a 730 kb long region containing the transcription factor-encoding genes SUPT3H and RUNX2, as well as the microRNA locus MIRN586. We also observed that this chromosomal segment coincides with pronounced linkage disequilibrium (LD), suggesting a relationship between TD and LD. -

Of Small Intestine Harboring Driver Gene Mutations: a Case Report and a Literature Review
1161 Case Report A rare multiple primary sarcomatoid carcinoma (SCA) of small intestine harboring driver gene mutations: a case report and a literature review Zhu Zhu1#, Xinyi Liu2#, Wenliang Li1, Zhengqi Wen1, Xiang Ji1, Ruize Zhou1, Xiaoyu Tuo3, Yaru Chen2, Xian Gong2, Guifeng Liu2, Yanqing Zhou2, Shifu Chen2, Lele Song2#^, Jian Huang1 1Department of Oncology, First Affiliated Hospital of Kunming Medical University, Kunming, China; 2HaploX Biotechnology, Shenzhen, China; 3Department of Pathology, First Affiliated Hospital of Kunming Medical University, Kunming, China #These authors contributed equally to this work. Correspondence to: Jian Huang. Department of Oncology, First Affiliated Hospital of Kunming Medical University, No. 295, Xichang Road, Kunming 560032, Yunnan Province, China. Email: [email protected]; Lele Song. HaploX Biotechnology, 8th floor, Auto Electric Power Building, Songpingshan Road, Nanshan District, Shenzhen 518057, Guangdong Province, China. Email: [email protected]. Abstract: Primary sarcomatoid carcinoma (SCA) is a type of rare tumor consisting of both malignant epithelial and mesenchymal components. Only 32 cases of SCA of the small bowel have been reported in the literature to date. Due to its rarity and complexity, this cancer has not been genetically studied and its diagnosis and treatment remain difficult. Here we report a 54-year-old male underwent emergency surgical resection in the small intestine due to severe obstruction and was diagnosed with multiple SCA based on postoperative pathological examination. Over 100 polypoid tumors scattered along his whole jejunum and proximal ileum. Chemotherapy (IFO+Epirubicin) was performed after surgery while the patient died two months after the surgery due to severe malnutrition. Whole-exome sequencing was performed for the tumor tissue with normal tissue as the control.