LESSON 2 DNA Barcoding and the Barcode of Life Database
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Order BERYCIFORMES ANOPLOGASTRIDAE Fangtooths (Ogrefish) by J.A
click for previous page 1178 Bony Fishes Order BERYCIFORMES ANOPLOGASTRIDAE Fangtooths (ogrefish) by J.A. Moore, Florida Atlantic University, USA iagnostic characters: Small (to about 160 mm standard length) beryciform fishes.Body short, deep, and Dcompressed, tapering to narrow peduncle. Head large (1/3 standard length). Eye smaller than snout length in adults, but larger than snout length in juveniles. Mouth very large and oblique, jaws extend be- hind eye in adults; 1 supramaxilla. Bands of villiform teeth in juveniles are replaced with large fangs on dentary and premaxilla in adults; vomer and palatines toothless. Deep sensory canals separated by ser- rated ridges; very large parietal and preopercular spines in juveniles of one species, all disappearing with age. Gill rakers as clusters of teeth on gill arch in adults (lath-like in juveniles). No true fin spines; single, long-based dorsal fin with 16 to 20 rays; anal fin very short-based with 7 to 9 soft rays; caudal fin emarginate; pectoral fins with 13 to 16 soft rays; pelvic fins with 7 soft rays. Scales small, non-overlapping, spinose, goblet-shaped in adults; lateral line an open groove partially bridged by scales; no enlarged ventral keel scutes. Colour: entirely dark brown or black in adults. Habitat, biology, and fisheries: Meso- to bathypelagic, at depths of 75 to 5 000 m. Carnivores, with juveniles feeding on mainly crustaceans and adults mainly on fishes. May sometimes swim in small groups. Uncommon deep-sea fishes of no commercial importance. Remarks: The family was revised recently by Kotlyar (1986) and contains 1 genus with 2 species throughout the tropical and temperate latitudes. -

Phylogenetic Relationships and Subgeneric Taxonomy of Toad�Headed Agamas Phrynocephalus (Reptilia, Squamata, Agamidae) As Determined by Mitochondrial DNA Sequencing E
ISSN 00124966, Doklady Biological Sciences, 2014, Vol. 455, pp. 119–124. © Pleiades Publishing, Ltd., 2014. Original Russian Text © E.N. Solovyeva, N.A. Poyarkov, E.A. Dunayev, R.A. Nazarov, V.S. Lebedev, A.A. Bannikova, 2014, published in Doklady Akademii Nauk, 2014, Vol. 455, No. 4, pp. 484–489. GENERAL BIOLOGY Phylogenetic Relationships and Subgeneric Taxonomy of ToadHeaded Agamas Phrynocephalus (Reptilia, Squamata, Agamidae) as Determined by Mitochondrial DNA Sequencing E. N. Solovyeva, N. A. Poyarkov, E. A. Dunayev, R. A. Nazarov, V. S. Lebedev, and A. A. Bannikova Presented by Academician Yu.Yu. Dgebuadze October 25, 2013 Received October 30, 2013 DOI: 10.1134/S0012496614020148 Toadheaded agamas (Phrynocephalus) is an essen Trapelus, and Stellagama) were used in molecular tial element of arid biotopes throughout the vast area genetic analysis. In total, 69 sequences from the Gen spanning the countries of Middle East and Central Bank were studied, 28 of which served as outgroups (the Asia. They constitute one of the most diverse genera of members of Agamidae, Chamaeleonidae, Iguanidae, the agama family (Agamidae), variously estimated to and Lacertidae). comprise 26 to 40 species [1]. The subgeneric Phryno The fragment sequences of the following four cephalus taxonomy is poorly studied: recent taxo mitochondrial DNA genes were used in phylogenetic nomic revision have been conducted without analysis analysis: the genes of subunit I of cytochrome c oxi of the entire genus diversity [1]; therefore, its phyloge dase (COI), of subunits II and IV of NADHdehydro netic position within Agamidae family remains genase (ND2 and ND4), and of cytochrome b (cyt b). -

Jolanta KEMPTER*, Maciej KIEŁPIŃSKI, Remigiusz PANICZ, and Sławomir KESZKA
ACTA ICHTHYOLOGICA ET PISCATORIA (2016) 46 (4): 287–291 DOI: 10.3750/AIP2016.46.4.02 MICROSATELLITE DNA-BASED GENETIC TRACEABILITY OF TWO POPULATIONS OF SPLENDID ALFONSINO, BERYX SPLENDENS (ACTINOPTERYGII: BERYCIFORMES: BERYCIDAE)—PROJECT CELFISH—PART 2 Jolanta KEMPTER*, Maciej KIEŁPIŃSKI, Remigiusz PANICZ, and Sławomir KESZKA Division of Aquaculture, West Pomeranian University of Technology, Szczecin, Kazimierza Krolewicza 4, 71-550 Szczecin, Poland Kempter J., Kiełpinski M., Panicz R., Keszka S. 2016. Microsatellite DNA-based genetic traceability of two populations of splendid alfonsino, Beryx splendens (Actinopterygii: Beryciformes: Berycidae)— Project CELFISH—Part 2. Acta Ichthyol. Piscat. 46 (4): 287–291. Background. The study is a contribution to Project CELFISH which involves genetic identifi cation of populations of fi sh species presenting a particular economic importance or having a potential to be used in the so-called commercial substitutions. The EU fi sh trade has been showing a distinct trend of more and more fi sh species previously unknown to consumers being placed on the market. Molecular assays have become the only way with which to verify the reliability of exporters. This paper is aimed at pinpointing genetic markers with which to label and differentiate between two populations of splendid alfonsino, Beryx splendens Lowe, 1834, a species highly attractive to consumers in Asia and Oceania due to the meat taste and low fat content. Material and methods. DNA was isolated from fragments of fi ns collected at local markets in Japan (MJ) (n = 10) and New Zealand (MNZ) (n = 18). The rhodopsin gene (RH1) fragment and 16 microsatellite DNA fragments (SSR) were analysed in all the individuals. -

Egyptian Tortoise (Testudo Kleinmanni)
EAZA Reptile Taxon Advisory Group Best Practice Guidelines for the Egyptian tortoise (Testudo kleinmanni) First edition, May 2019 Editors: Mark de Boer, Lotte Jansen & Job Stumpel EAZA Reptile TAG chair: Ivan Rehak, Prague Zoo. EAZA Best Practice Guidelines Egyptian tortoise (Testudo kleinmanni) EAZA Best Practice Guidelines disclaimer Copyright (May 2019) by EAZA Executive Office, Amsterdam. All rights reserved. No part of this publication may be reproduced in hard copy, machine-readable or other forms without advance written permission from the European Association of Zoos and Aquaria (EAZA). Members of the European Association of Zoos and Aquaria (EAZA) may copy this information for their own use as needed. The information contained in these EAZA Best Practice Guidelines has been obtained from numerous sources believed to be reliable. EAZA and the EAZA Reptile TAG make a diligent effort to provide a complete and accurate representation of the data in its reports, publications, and services. However, EAZA does not guarantee the accuracy, adequacy, or completeness of any information. EAZA disclaims all liability for errors or omissions that may exist and shall not be liable for any incidental, consequential, or other damages (whether resulting from negligence or otherwise) including, without limitation, exemplary damages or lost profits arising out of or in connection with the use of this publication. Because the technical information provided in the EAZA Best Practice Guidelines can easily be misread or misinterpreted unless properly analysed, EAZA strongly recommends that users of this information consult with the editors in all matters related to data analysis and interpretation. EAZA Preamble Right from the very beginning it has been the concern of EAZA and the EEPs to encourage and promote the highest possible standards for husbandry of zoo and aquarium animals. -

Order BERYCIFORMES ANOPLOGASTRIDAE Anoplogaster
click for previous page 2210 Bony Fishes Order BERYCIFORMES ANOPLOGASTRIDAE Fangtooths by J.R. Paxton iagnostic characters: Small (to 16 cm) Dberyciform fishes, body short, deep, and compressed. Head large, steep; deep mu- cous cavities on top of head separated by serrated crests; very large temporal and pre- opercular spines and smaller orbital (frontal) spine in juveniles of one species, all disap- pearing with age. Eyes smaller than snout length in adults (but larger than snout length in juveniles). Mouth very large, jaws extending far behind eye in adults; one supramaxilla. Teeth as large fangs in pre- maxilla and dentary; vomer and palatine toothless. Gill rakers as gill teeth in adults (elongate, lath-like in juveniles). No fin spines; dorsal fin long based, roughly in middle of body, with 16 to 20 rays; anal fin short-based, far posterior, with 7 to 9 rays; pelvic fin abdominal in juveniles, becoming subthoracic with age, with 7 rays; pectoral fin with 13 to 16 rays. Scales small, non-overlap- ping, spinose, cup-shaped in adults; lateral line an open groove partly covered by scales. No light organs. Total vertebrae 25 to 28. Colour: brown-black in adults. Habitat, biology, and fisheries: Meso- and bathypelagic. Distinctive caulolepis juvenile stage, with greatly enlarged head spines in one species. Feeding mode as carnivores on crustaceans as juveniles and on fishes as adults. Rare deepsea fishes of no commercial importance. Remarks: One genus with 2 species throughout the world ocean in tropical and temperate latitudes. The family was revised by Kotlyar (1986). Similar families occurring in the area Diretmidae: No fangs, jaw teeth small, in bands; anal fin with 18 to 24 rays. -

How Much Longer Will It Take?
How much longer will it take? A ten-year review of the implementation of United Nations General Assembly resolutions 61/105, 64/72 and 66/68 on the management of bottom fisheries in areas beyond national jurisdiction FULL REPORT – AUGUST 2016 DAVID SHALE/NATURE PICTURE LIBRARAY SHALE/NATURE DAVID Leiopathes sp., a deepwater black coral, has lifespans in excess of 4,200 years (Roark et al., 2009*), making it one of the oldest living organism on Earth. Specimen was located off the coast of Oahu, Hawaii, in ~400 m water depth. * Roark, E.B., Guilderson, T.P., Dunbar, R.B., Fallon, S.J., and Mucciarone, D.A., 2009. Extreme longevity in proteinaceous deep-sea corals. Proceedings of the National Academy of Sciences, 106: 520– 5208, doi: 10.1073/pnas.0810875106. © HAWAII UNDERSEA RESEARCH LABORATORY, TERRY KERBY AND MAXIMILIAN CREMER Contents Executive summary 03 1.0 Introduction 09 2.0 North Atlantic 10 2.1 Northeast Atlantic 10 2..2 Northwest Atlantic 21 3.0 South Atlantic 33 3.1 Southeast Atlantic 33 3.2 Southwest Atlantic and other non-RFMO areas 39 4.0 North Pacific 42 Citation: 5.0 South Pacific 49 Gianni, M., Fuller, S.D., Currie, D.E.J., Schleit, 6.0 Indian Ocean 60 K., Goldsworthy, L., Pike, B., Weeber, B., 7.0 Southern Ocean 66 Owen, S., Friedman, A. How much longer will it take? A ten-year 8.0. Mediterranean Sea 71 review of the implementation of United Annex 1. Acronyms 73 Nations General Assembly resolutions Annex 2. History of the UNGA negotiations 73 61/105, 64/72 and 66/68 on the management Annex 3. -

Updated Checklist of Marine Fishes (Chordata: Craniata) from Portugal and the Proposed Extension of the Portuguese Continental Shelf
European Journal of Taxonomy 73: 1-73 ISSN 2118-9773 http://dx.doi.org/10.5852/ejt.2014.73 www.europeanjournaloftaxonomy.eu 2014 · Carneiro M. et al. This work is licensed under a Creative Commons Attribution 3.0 License. Monograph urn:lsid:zoobank.org:pub:9A5F217D-8E7B-448A-9CAB-2CCC9CC6F857 Updated checklist of marine fishes (Chordata: Craniata) from Portugal and the proposed extension of the Portuguese continental shelf Miguel CARNEIRO1,5, Rogélia MARTINS2,6, Monica LANDI*,3,7 & Filipe O. COSTA4,8 1,2 DIV-RP (Modelling and Management Fishery Resources Division), Instituto Português do Mar e da Atmosfera, Av. Brasilia 1449-006 Lisboa, Portugal. E-mail: [email protected], [email protected] 3,4 CBMA (Centre of Molecular and Environmental Biology), Department of Biology, University of Minho, Campus de Gualtar, 4710-057 Braga, Portugal. E-mail: [email protected], [email protected] * corresponding author: [email protected] 5 urn:lsid:zoobank.org:author:90A98A50-327E-4648-9DCE-75709C7A2472 6 urn:lsid:zoobank.org:author:1EB6DE00-9E91-407C-B7C4-34F31F29FD88 7 urn:lsid:zoobank.org:author:6D3AC760-77F2-4CFA-B5C7-665CB07F4CEB 8 urn:lsid:zoobank.org:author:48E53CF3-71C8-403C-BECD-10B20B3C15B4 Abstract. The study of the Portuguese marine ichthyofauna has a long historical tradition, rooted back in the 18th Century. Here we present an annotated checklist of the marine fishes from Portuguese waters, including the area encompassed by the proposed extension of the Portuguese continental shelf and the Economic Exclusive Zone (EEZ). The list is based on historical literature records and taxon occurrence data obtained from natural history collections, together with new revisions and occurrences. -

Stellio' in Herpetology and a Comment on the Nomenclature and Taxonomy of Agamids of the Genus Agama (Sensu Lato) (Squamata: Sauria: Agamidae)
©Österreichische Gesellschaft für Herpetologie e.V., Wien, Austria, download unter www.biologiezentrum.at HERPETOZOA 8 (1/2): 3 - 9 Wien, 30. Juli 1995 A brief review of the origin and use of 'stellio' in herpetology and a comment on the nomenclature and taxonomy of agamids of the genus Agama (sensu lato) (Squamata: Sauria: Agamidae) Kurzübersicht über die Herkunft und Verwendung von "stellio" in der Herpetologie und Kommentar zur Nomenklatur und Taxonomie von Agamen, Gattung Agama (sensu lato) (Squamata: Sauria: Agamidae) KLAUS HENLE ABSTRACT The name 'stellio' has received a wide and variable application in herpetology. Its use antedates modern nomenclature. The name was applied mainly to various agamid and gekkonid species. In modern usage, confu- sion exists primarily with its use as a genus, i. e., Siellio. To solve this problem, STEJNEGER (1936) desig- nated Siellio saxatilis of LAURENTI, 1768 which is based on a figure in SEBA (1734) as the type species. This species is unidentifiable. In an unpublished thesis, MOODY (1980) split the genus Agama (s. 1.) into six genera. He overlooked STEJNEGER's (1936) designation and reused Stellio for the stellio-group of agamid lizards. Many authors followed MOODY (1980). Recently, some authors pointed out that Siellio is unavailable but did not fully dis- cuss the implications for agamid nomenclature. It is argued that a satisfactory nomenclature is difficult with current knowledge of agamid taxonomy. It is suggested to restrict Laudakia to L. tuberculata and to use Ploce- denna for the stellio-group (sensu stricto). KURZFASSUNG Der Name "stellio" sah eine breite und vielfältige Verwendung in der Herpetologie, die weit über die moderne Nomenklatur zurückreicht. -

Literature Cited in Lizards Natural History Database
Literature Cited in Lizards Natural History database Abdala, C. S., A. S. Quinteros, and R. E. Espinoza. 2008. Two new species of Liolaemus (Iguania: Liolaemidae) from the puna of northwestern Argentina. Herpetologica 64:458-471. Abdala, C. S., D. Baldo, R. A. Juárez, and R. E. Espinoza. 2016. The first parthenogenetic pleurodont Iguanian: a new all-female Liolaemus (Squamata: Liolaemidae) from western Argentina. Copeia 104:487-497. Abdala, C. S., J. C. Acosta, M. R. Cabrera, H. J. Villaviciencio, and J. Marinero. 2009. A new Andean Liolaemus of the L. montanus series (Squamata: Iguania: Liolaemidae) from western Argentina. South American Journal of Herpetology 4:91-102. Abdala, C. S., J. L. Acosta, J. C. Acosta, B. B. Alvarez, F. Arias, L. J. Avila, . S. M. Zalba. 2012. Categorización del estado de conservación de las lagartijas y anfisbenas de la República Argentina. Cuadernos de Herpetologia 26 (Suppl. 1):215-248. Abell, A. J. 1999. Male-female spacing patterns in the lizard, Sceloporus virgatus. Amphibia-Reptilia 20:185-194. Abts, M. L. 1987. Environment and variation in life history traits of the Chuckwalla, Sauromalus obesus. Ecological Monographs 57:215-232. Achaval, F., and A. Olmos. 2003. Anfibios y reptiles del Uruguay. Montevideo, Uruguay: Facultad de Ciencias. Achaval, F., and A. Olmos. 2007. Anfibio y reptiles del Uruguay, 3rd edn. Montevideo, Uruguay: Serie Fauna 1. Ackermann, T. 2006. Schreibers Glatkopfleguan Leiocephalus schreibersii. Munich, Germany: Natur und Tier. Ackley, J. W., P. J. Muelleman, R. E. Carter, R. W. Henderson, and R. Powell. 2009. A rapid assessment of herpetofaunal diversity in variously altered habitats on Dominica. -

Deep‐Time Convergent Evolution in Animal Communication Presented
Received: 18 November 2020 | Revised: 15 April 2021 | Accepted: 19 April 2021 DOI: 10.1111/ele.13773 LETTER Deep- time convergent evolution in animal communication presented by shared adaptations for coping with noise in lizards and other animals Terry J. Ord1 | Danielle A. Klomp1 | Thomas C. Summers1 | Arvin Diesmos2 | Norhayati Ahmad3 | Indraneil Das4 1Evolution & Ecology Research Centre Abstract and the School of Biological, Earth and Environmental Sciences, University of New Convergence in communication appears rare compared with other forms of ad- South Wales, Sydney, NSW, Australia aptation. This is puzzling, given communication is acutely dependent on the envi- 2Herpetology Section, Zoology Division, National Museum of the Philippines, ronment and expected to converge in form when animals communicate in similar Manila, Philippines habitats. We uncover deep- time convergence in territorial communication between 3Department of Biological Sciences and Biotechnology, Universiti Kebangsaan two groups of tropical lizards separated by over 140 million years of evolution: Malaysia, Bangi, Malaysia the Southeast Asian Draco and Caribbean Anolis. These groups have repeatedly 4Institute of Biodiversity and Environmental Conservation, Universiti converged in multiple aspects of display along common environmental gradients. Malaysia Sarawak, Kota Samarahan, Malaysia Robot playbacks to free- ranging lizards confirmed that the most prominent con- vergence in display is adaptive, as it improves signal detection. We then provide Correspondence Terry J. Ord, Evolution & Ecology evidence from a sample of the literature to further show that convergent adap- Research Centre and the School of Biological, Earth and Environmental tation among highly divergent animal groups is almost certainly widespread in Sciences, University of New South Wales, nature. -

Modelling the Distribution of Alfonsino, Beryx Splendens, Over The
Abstract.- Commercial and Modelling the distribution of scientificbottom longline catches of alfonsino, Beryx splendens, from alfonsino, Beryx splendens, over seamounts off New Caledonia were sampled to study length-frequency distributions. A total of 14,674 fish the seamounts of New Caledonia were measured. CPUE of Beryx splendens on two seamounts is mod- Patrick Lehodey elled in terms of length and depth. The data show that mean length in- Paul Marchal creases with depth; this is well de- scribed by a bivariate normal model Rene Grandperrin that estimates catch for a given sea- Centre ORSTOM, BP A5, Noumea, New Caledonia mount. In addition, the data show I that mean length also varies with the depth of the top of seamounts; this is described by a recursive model that is designed to predict ap- proximate catch for any seamount. A bottom longline fishery operated recursive model predicts catch on The limitations of both models are on the seamounts of the Exclusive any seamount. discussed, particularly with regard Economic Zone (EEZ) of New Cale- to temporal variation. donia from February 1988 to July 1991.l Three vessels were involved Material and methods but only one vessel was operated at any given time. The fishing effort, Data which totalled 4,691,635 hooks, fo- Alfonsino were captured with long- cused on five seamounts (B, C, D, line gear (Fig. 2). The main line, J, and K) whose summits are lo- averaging 4,000 m, was held on the cated at depths ranging from 500 1). bottom by means of terminal an- to 750 m (Fig. -
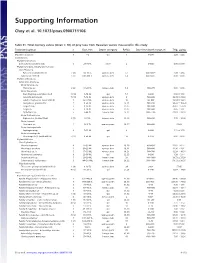
Supporting Information
Supporting Information Choy et al. 10.1073/pnas.0900711106 Table S1. Total mercury values (mean ؎ SD) of prey taxa from Hawaiian waters measured in this study Taxonomic group n Size, mm Depth category Ref(s). Day-time depth range, m THg, g/kg Mixed Zooplankton 5 1–2 epi 1 0–200 2.26 Ϯ 3.23 Invertebrates Phylum Ctenophora Ctenophores (unidentified) 3 20–30 TL other 2 0–600 0.00 Ϯ 0.00 Phylum Chordata, Subphylum Tunicata Class Thaliacea Pyrosomes (unidentified) 2 (8) 14–36 TL upmeso.dvm 2, 3 400–600ϩ 3.49 Ϯ 4.94 Salps (unidentified) 3 (7) 200–400 TL upmeso.dvm 2, 4 400–600ϩ 0.00 Ϯ 0.00 Phylum Arthropoda Subphylum Crustacea Order Amphipoda Phronima sp. 2 (6) 17–23 TL lomeso.dvm 5, 6 400–975 0.00 Ϯ 0.00 Order Decapoda Crab Megalopae (unidentified) 3 (14) 3–14 CL epi 7, 8 0–200 0.94 Ϯ 1.63 Janicella spinacauda 7 (10) 7–16 CL upmeso.dvm 9 500–600 30.39 Ϯ 23.82 Lobster Phyllosoma (unidentified) 5 42–67 CL upmeso.dvm 10 80–400 18.54 Ϯ 13.61 Oplophorus gracilirostris 5 9–20 CL upmeso.dvm 9, 11 500–650 90.23 Ϯ 103.20 Sergestes sp. 5 8–25 CL upmeso.dvm 12, 13 200–600 45.61 Ϯ 51.29 Sergia sp. 5 6–10 CL upmeso.dvm 12, 13 300–600 0.45 Ϯ 1.01 Systellapsis sp. 5 5–44 CL lomeso.dvm 9, 12 600–1100 22.63 Ϯ 38.18 Order Euphausiacea Euphausiids (unidentified) 2 (7) 5–7 CL upmeso.dvm 14, 15 400–600 7.72 Ϯ 10.92 Order Isopoda Anuropus sp.