Genetic Diversity in Tunisian Crataegus Azarolus L. Var. Aronia L
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
Seeds and Plants Imported
Issued November 9,1915. U. S. DEPARTMENT OF AGRICULTURE. BUREAU OF PLANT INDUSTRY. WILLIAM A. TAYLOR, Chief of Bureau. INVENTORY SEEDS AND PLANTS IMPORTED OFFICE OF FOREIGN SEED AND PLANT INTRODUCTION DURING THE PERIOD FROM APRIL 1 " " TO JUNE 30,1913. " , r (No. 35; Nos. 35136 TO 3566^..,--"•-****"*"' WASHINGTON: GOVERNMENT PRINTING OFFICE. 1915. Issued November 9,1915. U. S. DEPARTMENT OF AGRICULTURE. BUREAU OF PLANT INDUSTRY. WILLIAM A. TAYLOR, Chief of Bureau. INVENTORY SEEDS AND PLANTS IMPORTED BY THE OFFICE OF FOREIGN SEED AND PLANT INTRODUCTION DURING THE PERIOD FROM APRIL 1 TO JUNE 30,1913. (No. 35; Nos. 35136 TO 35666.) WASHINGTON: GOVERNMENT PRINTING OFFICE. 1915. BUREAU OF PLANT INDUSTRY. Chief of Bureau, WILLIAM A. TAYLOR. Assistant Chief of Bureau, KARL F. KELLERMAN. Officer in Charge of Publications, J. E. ROCKWELL. Chief Clerk, JAMES E. JONES. FOREIGN SEED AND PLANT INTRODUCTION. SCIENTIFIC STAFF. David Fairchild, Agricultural Explorer in Charge. P. H. Dorsett, Plant Introducer, in Charge of Plant Introduction Field Stations. Peter Bisset, Plant Introducer, in Charge of Foreign Plant Distribution. Frank N. Meyer and Wilson Popenoe, Agricultural Explorers. H. C. Skeels, S. C. Stuntz, and R. A. Young, Botanical Assistants. Allen M. Groves, Nathan Menderson, and Glen P. Van Eseltine, Assistants. Robert L. Beagles, Superintendent, Plant Introduction Field Station, Chico, Cal. Edward Simmonds, Superintendent, Subtropical Plant Introduction Field Station, Miami, Fla. John M. Rankin, Superintendent, Yarrow Plant Introduction Field Station, Rockville, Md. E. R. Johnston, Assistant in Charge, Plant Introduction Field Station, Brooksville, Fla. Edward Goucher and H. Klopfer, Plant Propagators. Collaborators: Aaron Aaronsohn, Director, Jewish Agricultural Experimental Station, Haifa, Palestine; Thomas W. -

What Is a Tree in the Mediterranean Basin Hotspot? a Critical Analysis
Médail et al. Forest Ecosystems (2019) 6:17 https://doi.org/10.1186/s40663-019-0170-6 RESEARCH Open Access What is a tree in the Mediterranean Basin hotspot? A critical analysis Frédéric Médail1* , Anne-Christine Monnet1, Daniel Pavon1, Toni Nikolic2, Panayotis Dimopoulos3, Gianluigi Bacchetta4, Juan Arroyo5, Zoltán Barina6, Marwan Cheikh Albassatneh7, Gianniantonio Domina8, Bruno Fady9, Vlado Matevski10, Stephen Mifsud11 and Agathe Leriche1 Abstract Background: Tree species represent 20% of the vascular plant species worldwide and they play a crucial role in the global functioning of the biosphere. The Mediterranean Basin is one of the 36 world biodiversity hotspots, and it is estimated that forests covered 82% of the landscape before the first human impacts, thousands of years ago. However, the spatial distribution of the Mediterranean biodiversity is still imperfectly known, and a focus on tree species constitutes a key issue for understanding forest functioning and develop conservation strategies. Methods: We provide the first comprehensive checklist of all native tree taxa (species and subspecies) present in the Mediterranean-European region (from Portugal to Cyprus). We identified some cases of woody species difficult to categorize as trees that we further called “cryptic trees”. We collected the occurrences of tree taxa by “administrative regions”, i.e. country or large island, and by biogeographical provinces. We studied the species-area relationship, and evaluated the conservation issues for threatened taxa following IUCN criteria. Results: We identified 245 tree taxa that included 210 species and 35 subspecies, belonging to 33 families and 64 genera. It included 46 endemic tree taxa (30 species and 16 subspecies), mainly distributed within a single biogeographical unit. -

Towards an Updated Checklist of the Libyan Flora
Towards an updated checklist of the Libyan flora Article Published Version Creative Commons: Attribution 3.0 (CC-BY) Open access Gawhari, A. M. H., Jury, S. L. and Culham, A. (2018) Towards an updated checklist of the Libyan flora. Phytotaxa, 338 (1). pp. 1-16. ISSN 1179-3155 doi: https://doi.org/10.11646/phytotaxa.338.1.1 Available at http://centaur.reading.ac.uk/76559/ It is advisable to refer to the publisher’s version if you intend to cite from the work. See Guidance on citing . Published version at: http://dx.doi.org/10.11646/phytotaxa.338.1.1 Identification Number/DOI: https://doi.org/10.11646/phytotaxa.338.1.1 <https://doi.org/10.11646/phytotaxa.338.1.1> Publisher: Magnolia Press All outputs in CentAUR are protected by Intellectual Property Rights law, including copyright law. Copyright and IPR is retained by the creators or other copyright holders. Terms and conditions for use of this material are defined in the End User Agreement . www.reading.ac.uk/centaur CentAUR Central Archive at the University of Reading Reading’s research outputs online Phytotaxa 338 (1): 001–016 ISSN 1179-3155 (print edition) http://www.mapress.com/j/pt/ PHYTOTAXA Copyright © 2018 Magnolia Press Article ISSN 1179-3163 (online edition) https://doi.org/10.11646/phytotaxa.338.1.1 Towards an updated checklist of the Libyan flora AHMED M. H. GAWHARI1, 2, STEPHEN L. JURY 2 & ALASTAIR CULHAM 2 1 Botany Department, Cyrenaica Herbarium, Faculty of Sciences, University of Benghazi, Benghazi, Libya E-mail: [email protected] 2 University of Reading Herbarium, The Harborne Building, School of Biological Sciences, University of Reading, Whiteknights, Read- ing, RG6 6AS, U.K. -

Effects of Methanolic Extract of Crataegus Azarolus L
Available online on www.ijppr.com International Journal of Pharmacognosy and Phytochemical Research 2016; 8(6); 968-976 ISSN: 0975-4873 Research Article Effects of Methanolic Extract of Crataegus azarolus L. on Plasma Antioxidant Capacity and Biomarkers of Oxidative Stress in Liver and Kidney of Rats 1 1* 1 2 1 1 2 Bouaziz A , Khennouf S , Djidel S , Baghiani A , Dahamna S , Amira S , Arrar L 1Laboratory of Phytotherapy Applied to Chronic Diseases, Department of Biology and Animal Physiology, Facult Nature and Life Sciences, University Ferhat Abbas, Setif 1, 19000, Algeria. 2Laboratory of Applied Biochemistry, Department of Biochemistry, Faculty of Nature and Life Sciences, University Ferhat Abbas, Setif 1, 19000, Algeria. Available Online: 9th June, 2016 ABSTRACT The aim of the present study was to investigate the effect of methanolic extract of Crataegus azarolus (MECA) on the plasma antioxidant capacity and oxidative stress markers in liver and kidney of rats. Albino rats were categorized into four groups. The first group served as the normal control, the second and the third groups were treated with MECA at dose of 100 and 200 mg/kg, respectively. Fourth group (positive group) was treated with 100 mg/kg of vitamin C for 14 consecutive days. The levels of protein, malondialdehyde (MDA) and reduced glutathione (GSH) and catalase (CAT) activity were assayed in liver and kidney homogenates using standard procedures. The results showed that MECA (100 and 200 mg/kg) caused significant elevation of plasma antioxidant capacity using DPPH radical scavenging activity, whereas only the dose of 200 mg/kg was found to increase significantly the plasma reducing power. -

Collection and Assessment of Traditional Medicinal Plants Used By
J HerbMed Pharmacol. 2016; 5(2): 54-60. Journal of HerbMed Pharmacology Journal homepage: http://www.herbmedpharmacol.com Collection and assessment of traditional medicinal plants used by the indigenous people of Dastena in Iran Habib-allah Mohammadi1*, Seyed-Ebrahim Sajjadi1, Mostafa Noroozi2, Mahmoud Mirhosseini3 1Department of Pharmacognosy, School of Pharmacy, Isfahan University of Medical Sciences, Isfahan, Iran 2Isfahan Center for Research of Agricultural Science and Natural Resources, Isfahan, Iran 3Medical Plants Research Center, Shahrekord University of Medical Sciences, Shahrekord, Iran A R T I C L E I N F O A B S T R A C T Article Type: Introduction: Nowadays, traditional and herbal medicines have attracted the attention of Original Article researchers all around the world and despite the development of synthetic drugs, demand for plant- based medicines is growing. The main reason for this growing trend is increasing public concerns Article History: about the adverse effects of synthetic medicines. Traditional medicine and ethnobotany are two Received: 7 November 2015 important issues that should be noted to achieve effective herbal medicines with considerable Accepted: 2 January 2016 therapeutic effects. Traditional medicine is based on experience of people over centuries and ethno-botany is based on recognition of the native plants. Iran has very high plant diversity because of its different climate, ecosystems and soil conditions. Regarding increasing demand for Keywords: medicinal plants, this study aimed to collect some native plant varieties growing in Dastena and to Herbs review some of local and folk application of these plants. Traditional uses Methods: In the present study, the plant species were collected during two consecutive years Ethnobotany (2013-2014) and systematically identified. -

Nutritional, Antioxidative, and Antimicrobial Analysis of The
ORIGINAL RESEARCH Nutritional, antioxidative, and antimicrobial analysis of the Mediterranean hackberry (Celtis australis L.) Ajda Ota1, Ana Miklavcˇicˇ Višnjevec2, Rajko Vidrih1, Željko Prgomet3, Marijan Necˇemer4, Janez Hribar1, Nina Gunde Cimerman1,5, Sonja Smole Možina1, Milena Bucˇar-Miklavcˇicˇ2 & Nataša Poklar Ulrih1,5 1Biotechnical Faculty, University of Ljubljana, Jamnikarjeva 101, Ljubljana SI-1000, Slovenia 2Science and Research Centre of Koper, University of Primorska, Zelena ulica 8, Izola SI-6310, Slovenia 3Agricultural Department in Poreč, University of Rijeka, Carla Huguesa 6, Porecˇ, Croatia 4Institute Jožef Stefan, Jamova cesta 39, Ljubljana SI-1000, Slovenia 5Centre of Excellence for Integrated Approaches in Chemistry and Biology of Proteins (CIPKeBiP), Jamova cesta 39, Ljubljana SI-1000, Slovenia Keywords Abstract Antimicrobial activity, Celtis australis, nutritional analysis, phenols, triple- Celtis australis is a deciduous tree commonly known as Mediterranean hackberry quadrupole tandem mass spectrometry, or the European nettle tree. The fruit of hackberry are seldom used for nutri- ultrahigh-pressure liquid chromatography tional purposes. The nutritional and physicochemical properties of ripe hackberry fruit from Istria (Marasi village near Vrsar, Croatia) were determined, including Correspondence water, total fiber, protein, vitamin, mineral, and phenolic contents. This analysis Nataša Poklar Ulrih, Department of Food Science and Technology,Biotechnical Faculty, demonstrates that the hackberry fruit is a valuable source of dietary fiber, pro- University of Ljubljana, Jamnikarjeva 101, tein, and vitamins, and of pigments such as lutein, β-carotene, zeaxanthin, and SI-1000 Ljubljana, Slovenia. tocopherols. The seasonal differences associated with the different growth stages Tel: +386 1 320 3780; for the element composition, total phenolic content, and phenolic profile were Fax: +386 1 256 6296; also determined for hackberry mesocarp and leaves. -

Amaryllidaceae) in Iraq
Anales del Jardín Botánico de Madrid 74(1): e053 2017. ISSN: 0211-1322. doi: http://dx.doi.org/10.3989/ajbm.2451 On the genus Sternbergia (Amaryllidaceae) in Iraq Sami Youssef1, Ahmed Mahmood1 & Errol Vela2* 1 Department of Recreation and Ecotourism, College of Agriculture, University of Duhok, Sumail-Duhok 1063 BD, Kurdistan Region, Iraq 2 University of Montpellier, UMR AMAP (Botany and Modelisation of Plant Architecture), CIRAD TA A51/PS2, 34398 Montpellier cedex 5, France; [email protected] Abstract Resumen Youssef, S., Mahmood, A. & Vela, E. 2017. On the genus Sternbergia Youssef, S., Mahmood, A. & Vela, E. 2017. Sobre el género Sternbergia (Amaryllidaceae) in Iraq. Anales Jard. Bot. Madrid 74(1): e053. (Amaryllidaceae) en Iraq. Anales Jard. Bot. Madrid 74(1): e053. Sternbergia is a genus containing mostly remarkable autum flowering taxa Sternbergia contiene, sobre todo, extraordinarios táxones con floración oto- within Amaryllidaceae. Its distribution ranges from the Mediterranean ñal de las amarilidáceas. Su distribución abarca desde la región mediterránea, region through the Irano-Anatolian region to Caucasus and Central a través de la región irano-anatólica, hasta el Cáucaso y el Asia central. En la Asia. In Flora of Iraq, the information about the occurrence, habitat, and Flora de Iraq, la información sobre la presencia, el hábitat y la distribución de distribution of its species is outdated or incomplete. The main aim of sus especies está obsoleta o incompleta. El principal objetivo de este trabajo this study has been to contribute with new data from the field in order ha sido contribuir con datos nuevos tomados en el campo a actualizar su esta- to update its status in the Kurdistan Region. -

Types of Headache and Those Remedies in Traditional Persian Medicine
PHCOG REV. REVIEW ARTICLE Types of headache and those remedies in traditional persian medicine Mohammad M. Zarshenas1,2, Peyman Petramfar3, Ali Firoozabadi4, Mahmood Reza Moein5, Abdolali Mohagheghzadeh1,2 1Research Center for Traditional Medicine and History of Medicine, Shiraz University of Medical Sciences, 2Department of Traditional Pharmacy, School of Pharmacy, Shiraz University of Medical Sciences, 3Department of Neurology, School of Medicine, Shiraz University of Medical Sciences, 4Department of Psychiatry, School of Medicine, Shiraz University of Medical Sciences, 5Department of Pharmacognosy, School of Pharmacy, Shiraz University of Medical Sciences, Shiraz, Iran Submitted: 27-06-2012 Revised: 29-12-2012 Published: 01-06-2013 ABSTRACT The history of headache, as a common neurological complication, goes back to almost 9000 years ago. Many ancient civilizations present references to headaches and the coherent treatment strategies. Accordingly, several documents comprising headache complications embodying precise medical information stem from Traditional Persian Medicine (TPM) that can provide useful opportunities for more comprehensive treatment. We conducted a survey on headache through original important pharmacopeias and other important medical manuscripts of TPM which were written during 9th to 19th centuries and have derived all headache categories and herbal remedies. An extensive search of scientific data banks, such as Medline and Scopus, has also been exercised to find results relating to the anti‑inflammatory, anti‑nociceptive, and analgesic effects of denoted medicinal herbs. The concept of headache and treatments in TPM covers over 20 various types of headache and more than 160 different medicinal plants administered for oral, topical, and nasal application according to 1000 years of the subject documents. Nearly, 60% of remarked medicinal herbs have related anti‑inflammatory or analgesic effects and some current headache types have similarities and conformities to those of traditional types. -

Crataegus Spp.) Fruits Species for Potential Use in Food Applications
foods Article Physicochemical Characterization, Antioxidant Activity, and Phenolic Compounds of Hawthorn (Crataegus spp.) Fruits Species for Potential Use in Food Applications Abolfazl Alirezalu 1 , Nima Ahmadi 2,*, Peyman Salehi 3, Ali Sonboli 3, Kazem Alirezalu 4, Amin Mousavi Khaneghah 5, Francisco J. Barba 6 , Paulo E.S. Munekata 7 and Jose M. Lorenzo 7,* 1 Department of Horticultural Sciences, Faculty of Agriculture, Urmia University, Urmia 5756151818, Iran; [email protected] 2 Department of Horticultural Sciences, Faculty of Agriculture, Tarbiat Modares University, Tehran 1411713116, Iran 3 Medicinal Plants and Drugs Research Institute, Shahid Beheshti University, Tehran 1983969411, Iran; [email protected] (P.S.); [email protected] (A.S.) 4 Department of Food Science and Technology, Ahar Faculty of Agriculture and Natural Resources, University of Tabriz, Tabriz, Iran; [email protected] 5 Department of Food Science, Faculty of Food Engineering, University of Campinas (UNICAMP), Campinas, 13083-862 São Paulo, Brazil; [email protected] 6 Nutrition and Food Science Area, Preventive Medicine and Public Health, Food Science, Toxicology and Forensic Medicine Department, Faculty of Pharmacy, Universitat de València, Avda.Vicent Andrés Estellés, s/n, 46100 Burjassot, València, Spain; [email protected] 7 Centro Tecnológico de la Carne de Galicia, rúa Galicia n◦ 4, Parque Tecnológico de Galicia, San Cibrao das Viñas, 32900 Ourense, Spain; [email protected] * Correspondence: [email protected] (N.A.); [email protected] (J.M.L.); Tel.: +98 21 48292088 (N.A.); +34-988548277 (J.M.L.) Received: 11 March 2020; Accepted: 1 April 2020; Published: 4 April 2020 Abstract: Hawthorn belongs to the Crataegus genus of the Rosaceae family and is an important medicinal plant. -
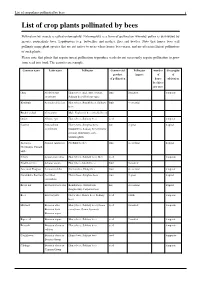
List of Crop Plants Pollinated by Bees 1 List of Crop Plants Pollinated by Bees
List of crop plants pollinated by bees 1 List of crop plants pollinated by bees Pollination by insects is called entomophily. Entomophily is a form of pollination whereby pollen is distributed by insects, particularly bees, Lepidoptera (e.g. butterflies and moths), flies and beetles. Note that honey bees will pollinate many plant species that are not native to areas where honey bees occur, and are often inefficient pollinators of such plants. Please note that plants that require insect pollination to produce seeds do not necessarily require pollination to grow from seed into food. The carrot is an example. Common name Latin name Pollinator Commercial Pollinator number Geography product impact of of of pollination honey cultivation bee hives per acre Okra Abelmoschus Honey bees (incl. Apis cerana), fruit 2-modest temperate esculentus Solitary bees (Halictus spp.) Kiwifruit Actinidia deliciosa Honey bees, Bumblebees, Solitary fruit 4-essential bees Bucket orchid Coryanthes Male Euglossini bees (Orchid bees) Onion Allium cepa Honey bees, Solitary bees seed temperate Cashew Anacardium Honey bees, Stingless bees, nut 3-great tropical occidentale bumblebees, Solitary bees (Centris tarsata), Butterflies, flies, hummingbirds Atemoya, Annona squamosa Nitidulid beetles fruit 4-essential tropical Cherimoya, Custard apple Celery Apium graveolens Honey bees, Solitary bees, flies seed temperate Strawberry tree Arbutus unedo Honey bees, bumblebees fruit 2-modest American Pawpaw Asimina triloba Carrion flies, Dung flies fruit 4-essential temperate Carambola, -

Crataegus Spp.) from Different Regions of Iran
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Archivio della ricerca - Università degli studi di Napoli Federico II International Journal of Food Properties ISSN: 1094-2912 (Print) 1532-2386 (Online) Journal homepage: http://www.tandfonline.com/loi/ljfp20 Flavonoids profile and antioxidant activity in flowers and leaves of hawthorn species (Crataegus spp.) from different regions of Iran Abolfazl Alirezalu, Peyman Salehi, Nima Ahmadi, Ali Sonboli, Serena Aceto, Hamid Hatami Maleki & Mahdi Ayyari To cite this article: Abolfazl Alirezalu, Peyman Salehi, Nima Ahmadi, Ali Sonboli, Serena Aceto, Hamid Hatami Maleki & Mahdi Ayyari (2018) Flavonoids profile and antioxidant activity in flowers and leaves of hawthorn species (Crataegus spp.) from different regions of Iran, International Journal of Food Properties, 21:1, 452-470, DOI: 10.1080/10942912.2018.1446146 To link to this article: https://doi.org/10.1080/10942912.2018.1446146 © 2018 Abolfazl Alirezalu, Peyman Salehi, Nima Ahmadi, Ali Sonboli, Serena Aceto, Hamid Hatami Maleki, and Mahdi Ayyari. Published with license by Taylor & Francis. Accepted author version posted online: 11 Apr 2018. Published online: 11 Apr 2018. Submit your article to this journal Article views: 126 View related articles View Crossmark data Full Terms & Conditions of access and use can be found at http://www.tandfonline.com/action/journalInformation?journalCode=ljfp20 INTERNATIONAL JOURNAL OF FOOD PROPERTIES 2018, VOL. 21, NO. 1, 452–470 https://doi.org/10.1080/10942912.2018.1446146 -

Polyphenolic Composition of Crataegus Monogyna Jacq.: from Chemistry to Medical Applications
Nutrients 2015, 7, 7708-7728; doi:10.3390/nu7095361 OPEN ACCESS nutrients ISSN 2072-6643 www.mdpi.com/journal/nutrients Review Polyphenolic Composition of Crataegus monogyna Jacq.: From Chemistry to Medical Applications Seyed Fazel Nabavi 1, Solomon Habtemariam 2, Touqeer Ahmed 3, Antoni Sureda 4, Maria Daglia 5,*, Eduardo Sobarzo-Sánchez 6 and Seyed Mohammad Nabavi 1,* 1 Applied Biotechnology Research Center, Baqiyatallah University of Medical Sciences, Tehran 1193653471, Iran; E-Mail: [email protected] 2 Pharmacognosy Research Laboratories, Medway School of Science, University of Greenwich, Chatham-Maritime, Kent ME4 4TB, UK; E-Mail: [email protected] 3 Neurobiology Laboratory, Atta-ur-Rahman School of Applied Biosciences, National University of Sciences and Technology, Sector H-12, Islamabad 44000, Pakistan; E-Mail: [email protected] 4 Research Group on Community Nutrition and Oxidative Stress, University of Balearic Islands, and CIBEROBN (Physiopathology of Obesity and Nutrition), Palma de Mallorca E-07122, Spain; E-Mail: [email protected] 5 Department of Drug Sciences, Medicinal Chemistry and Pharmaceutical Technology Section, University of Pavia, Via Taramelli 12, Pavia 27100, Italy 6 Laboratory of Pharmaceutical Chemistry, Department of Organic Chemistry, Faculty of Pharmacy, University of Santiago de Compostela, Galicia 15782, Spain; E-Mail: [email protected] * Authors to whom correspondence should be addressed; E-Mails: [email protected] (M.D.); [email protected] (S.M.N.); Tel.: +39-0382-987388 (M.D.); +98-21-8861-7712 (S.M.N.); Fax: +39-0382-422975 (M.D.); +98-21-8861-7712 (S.M.N.). Received: 21 June 2015 / Accepted: 27 August 2015 / Published: 11 September 2015 Abstract: The abundance of scientific evidence has shown that many synthetic drugs can cause serious adverse effects in patients.