Epigenetic Control of Reprogramming and Cellular Differentiation
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Epigenetic Regulation of Muscle Stem Cell Expansion
Epigenetic regulation of muscle stem cell expansion INAUGURAL-DISSERTATION zur Erlangung des Doktorgrades der Naturwissenschaften - Doctor rerum naturalium - (Dr. rer. nat.) vorgelegt dem Fachbereich für Biologie und Chemie (FB 08) der Justus-Liebig-Universität Gießen eingereicht von Ting Zhang Gießen, 2015 Die vorliegende Arbeit wurde am Max-Planck-Institut für Herz- und Lungenforschung, W.G. Kerckhoff-Institut in Bad Nauheim angefertigt. Erstgutachter: Prof. Dr. Dr. Thomas Braun Abteilung Entwicklung und Umbau des Herzens Max-Planck-Institut für Herz- und Lungenforschung Ludwigstraße 43, 61231 Bad Nauheim Zweitgutachter: Prof. Dr. Lienhard Schmitz Biochemisches Institut Fachbereich Medizin Justus-Liebig-Universität Gießen Friedrichstrasse 24, 35392 Giessen Disputation am 10.Juni 2015 EIDESSTATTLICHE ERKLÄRUNG „Ich erkläre: Ich habe die vorgelegte Dissertation selbständig und ohne unerlaubte fremde Hilfe und nur mit den Hilfen angefertigt, die ich in der Dissertation angegeben habe. Alle Textstellen, die wörtlich oder sinngemäß aus veröffentlichten Schriften entnommen sind, und alle Angaben, die auf mündlichen Auskünften beruhen, sind als solche kenntlich gemacht. Bei den von mir durchgeführten und in der Dissertation erwähnten Untersuchungen habe ich die Grundsätze guter wissenschaftlicher Praxis, wie sie in der „Satzung der Justus-Liebig-Universität Gießen zur Sicherung guter wissenschaftlicher Praxis“ niedergelegt sind, eingehalten.“ Bad Nauheim, den ZUSAMMENFASSUNG ZUSAMMENFASSUNG Stammzellen der adulten Skelettmuskulatur, auch als -

Subcloning, Expression, and Enzymatic Study of PRMT5
Georgia State University ScholarWorks @ Georgia State University Biology Theses Department of Biology Summer 7-12-2010 Subcloning, Expression, and Enzymatic Study of PRMT5 Ran Guo Georgia State University Follow this and additional works at: https://scholarworks.gsu.edu/biology_theses Part of the Biology Commons Recommended Citation Guo, Ran, "Subcloning, Expression, and Enzymatic Study of PRMT5." Thesis, Georgia State University, 2010. https://scholarworks.gsu.edu/biology_theses/26 This Thesis is brought to you for free and open access by the Department of Biology at ScholarWorks @ Georgia State University. It has been accepted for inclusion in Biology Theses by an authorized administrator of ScholarWorks @ Georgia State University. For more information, please contact [email protected]. SUBCLONING, EXPRESSION, AND ENZYMATIC STUDY OF PRMT5 by RAN GUO Under the Direction of Yujun George Zheng ABSTRACT Protein arginine methyltransferases (PRMTs) mediate the transfer of methyl groups to arginine residues in histone and non-histone proteins. PRMT5 is an important member of PRMTs which symmetrically dimethylates arginine 8 in histone H3 (H3R8) and arginine 3 in histone H4 (H4R3). PRMT5 was reported to inhibit some tumor suppressors in leukemia and lymphoma cells and regulate p53 gene, through affecting the promoter of p53. Through methylation of H4R3, PRMT5 can recruit DNA-methyltransferase 3A (DNMT3A) which regulates gene transcription. All the above suggest that PRMT5 has an important function of suppressing cell apoptosis and is a potential anticancer target. Currently, the enzymatic activities of PRMT5 are not clearly understood. In our study, we improved the protein expression methodology and greatly enhanced the yield and quality of the recombinant PRMT5. -

UNIVERSITY of CALIFORNIA, IRVINE Combinatorial Regulation By
UNIVERSITY OF CALIFORNIA, IRVINE Combinatorial regulation by maternal transcription factors during activation of the endoderm gene regulatory network DISSERTATION submitted in partial satisfaction of the requirements for the degree of DOCTOR OF PHILOSOPHY in Biological Sciences by Kitt D. Paraiso Dissertation Committee: Professor Ken W.Y. Cho, Chair Associate Professor Olivier Cinquin Professor Thomas Schilling 2018 Chapter 4 © 2017 Elsevier Ltd. © 2018 Kitt D. Paraiso DEDICATION To the incredibly intelligent and talented people, who in one way or another, helped complete this thesis. ii TABLE OF CONTENTS Page LIST OF FIGURES vii LIST OF TABLES ix LIST OF ABBREVIATIONS X ACKNOWLEDGEMENTS xi CURRICULUM VITAE xii ABSTRACT OF THE DISSERTATION xiv CHAPTER 1: Maternal transcription factors during early endoderm formation in 1 Xenopus Transcription factors co-regulate in a cell type-specific manner 2 Otx1 is expressed in a variety of cell lineages 4 Maternal otx1 in the endodermal conteXt 5 Establishment of enhancers by maternal transcription factors 9 Uncovering the endodermal gene regulatory network 12 Zygotic genome activation and temporal control of gene eXpression 14 The role of maternal transcription factors in early development 18 References 19 CHAPTER 2: Assembly of maternal transcription factors initiates the emergence 26 of tissue-specific zygotic cis-regulatory regions Introduction 28 Identification of maternal vegetally-localized transcription factors 31 Vegt and OtX1 combinatorially regulate the endodermal 33 transcriptome iii -

Reversible Histone Glycation Is Associated with Disease-Related Changes in Chromatin Architecture
ARTICLE https://doi.org/10.1038/s41467-019-09192-z OPEN Reversible histone glycation is associated with disease-related changes in chromatin architecture Qingfei Zheng 1, Nathaniel D. Omans1,2, Rachel Leicher3,4, Adewola Osunsade 1,4, Albert S. Agustinus 1, Efrat Finkin-Groner1, Hannah D’Ambrosio1, Bo Liu5, Sarat Chandarlapaty 5, Shixin Liu3 & Yael David1,4,6,7 Cellular proteins continuously undergo non-enzymatic covalent modifications (NECMs) that accumulate under normal physiological conditions and are stimulated by changes in the 1234567890():,; cellular microenvironment. Glycation, the hallmark of diabetes, is a prevalent NECM asso- ciated with an array of pathologies. Histone proteins are particularly susceptible to NECMs due to their long half-lives and nucleophilic disordered tails that undergo extensive regulatory modifications; however, histone NECMs remain poorly understood. Here we perform a detailed analysis of histone glycation in vitro and in vivo and find it has global ramifications on histone enzymatic PTMs, the assembly and stability of nucleosomes, and chromatin archi- tecture. Importantly, we identify a physiologic regulation mechanism, the enzyme DJ-1, which functions as a potent histone deglycase. Finally, we detect intense histone glycation and DJ-1 overexpression in breast cancer tumors. Collectively, our results suggest an additional mechanism for cellular metabolic damage through epigenetic perturbation, with implications in pathogenesis. 1 Chemical Biology Program, Sloan Kettering Institute, Memorial Sloan Kettering Cancer Center, New York, NY 10065, USA. 2 Tri-Institutional Training Program in Computational Biology and Medicine, New York, NY 10065, USA. 3 Laboratory of Nanoscale Biophysics and Biochemistry, Rockefeller University, New York, NY 10065, USA. 4 Tri-institutional PhD Program in Chemical Biology, New York, NY 10065, USA. -

PRMT5 Modulates the Metabolic Response to Fasting Signals
PRMT5 modulates the metabolic response to fasting signals Wen-Wei Tsaia, Sherry Niessenb, Naomi Goebela, John R. Yates IIIb, Ernesto Guccionec,d, and Marc Montminya,1 aThe Clayton Foundation Laboratories for Peptide Biology, Salk Institute, La Jolla, CA 92037; bDepartment of Chemical Physiology, The Scripps Research Institute, La Jolla, CA 92037; cInstitute of Molecular and Cell Biology, Proteos, Singapore 138673; dDepartment of Biochemistry, Yong Loo Lin School of Medicine, National University of Singapore, Singapore 119074 Contributed by Marc Montminy, March 11, 2013 (sent for review February 20, 2013) Under fasting conditions, increases in circulating glucagon maintain sites. The results provide a mechanism to explain how latent cy- glucose balance by promoting hepatic gluconeogenesis. Triggering toplasmic regulators such as CRTC2 may contribute to signaling of the cAMP pathway stimulates gluconeogenic gene expression in the nucleus through their association with chromatin mod- through the PKA-mediated phosphorylation of the cAMP response ifying enzymes. element binding (CREB) protein and via the dephosphorylation of the latent cytoplasmic CREB regulated transcriptional coactivator 2 Results (CRTC2). CREB and CRTC2 activities are increased in insulin resis- In mass spectroscopy studies using epitope-tagged CRTC2 to tance, in which they promote hyperglycemia because of constitu- identify relevant interacting proteins, we recovered the protein tive induction of the gluconeogenic program. The extent to which arginine methyltransferase 5 (PRMT5) (Fig. S1 A and B). We CREB and CRTC2 are coordinately up-regulated in response to glu- confirmed the CRTC2:PRMT5 interaction in coimmunopreci- cagon, however, remains unclear. Here we show that, following its pitation studies with epitope-tagged proteins (Fig. -

H3r8me2(Asym) Polyclonal Antibody - Classic
H3R8me2(asym) polyclonal antibody - Classic Cat. No. C15410286 Specificity: Human, mouse, C. elegans, rat, chicken, Type: Polyclonal Xenopus, Drosophila, plant Source: Rabbit Purity: Affinity purified Lot #: 001 Storage: Store at -20°C; for long storage, store at -80°C. Avoid multiple freeze-thaw cycles. Size: 50 µg Precautions: This product is for research use only. Not for Concentration: 0.84 µg/µl use in diagnostic or therapeutic procedures. Applications Suggested dilution Results ChIP 2-5 µg/million cells Figure 1 Immunohistochemistry 1:100 IF 1:200 Figure 2 Western blot 1:500 Figure 3 Target description Chromatin is the arrangement of DNA and proteins in which chromosomes are formed. Correspondingly, chromatin is formed from nucleosomes, which are comprised of a set of four histone proteins (H2A, H2B, H3, H4) wrapped with DNA. Chromatin is a very dynamic structure in which numerous post-translational modifications work together to activate or repress the availability of DNA to be copied, transcribed, or repaired. These marks decide which DNA will be open and commonly active (euchromatin) or tightly wound to prevent access and activation (heterochromatin). Common histone modifications include methylation of lysine and arginine, acetylation of lysine, phosphorylation of threonine and serine, and sumoylation, biotinylation, and ubiquitylation of lysine. In particular, dimethylation of H3 Arg8 (H3 R8Me2) is known as a mark of transcriptional repression. The protein arginine methyltransferases PRMT5 and PRMT2 can both methylate Arg8, with PRMT2 specifically methylating in an asymmetric manner. In addition, asymmetric dimethylation of Arg8 inhibits H3K9 methylation by G9a, but symmetric Arg8Me2 does not. 1 Results Figure 1. -

Symmetric-Di-Methyl-Histone H3 (Arg8) Rabbit
Symmetric -Di-methyl-Histone H3 (Arg8) rabbit pAb ( H3R8me2(sy) pAb) www.ptm-biolab.com.cn Cat#: PTM-672 [email protected] Species: Rabbit 0571-2883 3567 Size: 100 μl Form: supplied in liquid form Recommend Species Swissprot Molecular Application Immunogen dilution Reactivity ID Weight H3R8me2(sy) WB, Dot, ELISA 1:2000 for WB H, M, R, Mk P68431 17 KDa -KLH **Species reactivity is determined by WB. Kept at -20oC after reconstituted. *** Anti-rabbit secondary antibodies must be used to detect this antibody. Source/Purification: This product is produced by immunizing rabbits with a synthetic di-methyl(sy) peptide corresponding to residues surrounding Arg8 of human histone H3. Antibodies are purified by protein A-conjugated agarose followed by di-methylated(sy) histone H3 (Arg8) peptide affinity chromatography. Recommended Applications: ELISA, Dot blot, Western blot. Recommended antibody dilution: WB: 1:2000 NOTE: For WB, incubate membrane with diluted antibody in 5% nonfat milk, 1 x TBS, 0.1% Tween- 20 for two hours at room temperature with gentle shaking. Prepare working dilution immediately before use. Use at an assay dependent concentration. Optimal dilutions/concentrations should be determined by the end user. Not yet tested in other applications Scientific Description: Proteins post-translational Modification (PTM) is an important reversible event controlling protein activity. The amino-terminal tails of core histones undergo multiple modifications in multiple sites, termed as “histone code” or “epigenetic code”. Lysine methylation or arginine methylation in core histones is a major determinant for the formation of active and inactive regions of the genome and therefore plays vital roles in multiple cellular events. -

H3r8me2(Asym) | Histone H3 (Asym-Dimethylated Arg8) Product Information
Product no AS16 3177 H3R8me2(asym) | Histone H3 (asym-dimethylated Arg8) Product information Immunogen KLH-conjugated synthetic peptide Host Rabbit Clonality Polyclonal Purity Affinity purified serum Format Liquid Quantity 50 µg Storage Store lyophilized/reconstituted at -20°C; once reconstituted make aliquots to avoid repeated freeze-thaw cycles. Please, remember to spin tubes briefly prior to opening them to avoid any losses that might occur from lyophilized material adhering to the cap or sides of the tubes. Additional information This antibody preparation is provided in 20 mM Potassium Phosphate pH 7.2, 150 mM NaCl, 0,01% sodium azide and 30% glycerol. Application information Recommended dilution 2 ug (ChIP), 1: 100 (IF), 1: 100 (IHC), 1:500 (WB) Expected | apparent 15 kDa MW Confirmed reactivity Caenorhabditis elegans, Human Predicted reactivity Chicken, Drosophila melanogaster, Mouse, Plant, Rat, Xenopus sp. Not reactive in No confirmed exceptions from predicted reactivity are currently known. application example Chromatin Immunoprecipitation using anti-H3R8me2(asym) antibodies. Chromatin from one million formaldehyde cross-linked Hela cells was used with 2 µg of H3R8me2(asym) and 20ul of magnetic beads per immunoprecipitation. A no antibody (No Ab) control was also used. Immunoprecipitated DNA was quantified using quantitative real-time PCR, and normalized to the input chromatin. Immunofluorescence using anti-H3R8me2(asym) antibodies. Tissue: HeLa cells. Fixation: 0.5% PFA. Primary antibody: used at a 1:100 dilution for 1 h at RT. Secondary antibody: FITC secondary antibody at 1:10 000 for 45 min at RT. Localization: H3R8me2(asym) is nuclear and chromosomal. Staining: H3R8me2(asym) is expressed in green and the nuclei and alpha-tubulin are counterstained with DAPI (blue) and Dylight® 594 (red). -
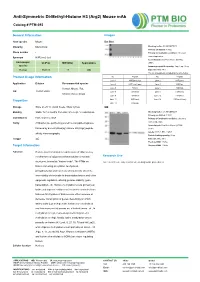
Anti-Symmetric Di-Methyl-Histone H3 (Arg2) Mouse Mab
Anti-Symmetric Di-Methyl-Histone H3 (Arg2) Mouse mAb Catalog # PTM-693 General Information Images Host species Mouse Dot Blot Clonality Monoclonal Blocking buffer: 5% NFDM/TBST Primary ab dilution: 1:1000 Clone number / Primary ab incubation condition: 2 hours at room temperature Synonym H3R2me2 (sy) Secondary ab: Goat Anti-Mouse IgG H&L Immunogen UniProt MW (kDa) Applications (HRP) species Immunogen peptide quantity: 1 ng, 4 ng, 16 ng Human P68431 17 WB Exposure time: 60 s The list of peptides is included in the table below: No. Peptide No. Peptide Product Usage Information Lane 1 H3R2me2 (sy) Lane 2 H3R2me1 Application Dilution Recommended species Lane 3 H3R2me2 (asy) Lane 4 H3K4ac Human, Mouse, Rat, Lane 5 H3K4cr Lane 6 H3K4hib WB 1:500-1:2000 Lane 7 H3K4me1 Lane 8 H3K4me2 Chlorocebus aethiops Lane 9 H3K4me3 Lane 10 H3R8me1 Lane 11 H3R8me2 Lane 12 H3R8me2 (asy) Properties Lane 13 H3R2un Storage Store at -20 °C. Avoid freeze / thaw cycles. WB Blocking buffer: 5% NFDM/TBST Stability Stable for 12 months from date of receipt / reconstitution Primary ab dilution: 1:2000 Constituents PBS, Glycerol, BSA Primary ab incubation condition: 2 hours at room temperature Purity Antibodies are purified by protein G-conjugated agarose Secondary ab: Goat Anti-Mouse IgG H&L followed by di-methylated(sy) histone H3 (Arg2) peptide (HRP) Lysate: MCF-7, BRL, COS-7 affinity chromatography. Protein loading quantity: 20 μg Isotype IgG Exposure time: 60 s Predicted MW: 17 kDa Target Information Observed MW: 17 kDa Function Histone post-translational modifications (PTMs) are key mechanisms of epigenetics that modulate chromatin Research Use structures, termed as “histone code”. -

Anti-H3r8me2(Asym) Antibody
FOR RESEARCH USE ONLY! 01/20 Anti-H3R8me2(asym) Antibody CATALOG NO.: A2047-100 (100 µl) BACKGROUND DESCRIPTION: Histones are basic nuclear proteins that are responsible for the nucleosome structure of the chromosomal fiber in eukaryotes. Nucleosomes consist of approximately 146 bp of DNA wrapped around a histone octamer composed of pairs of each of the four core histones (H2A, H2B, H3, and H4). The chromatin fiber is further compacted through the interaction of a linker histone, H1, with the DNA between the nucleosomes to form higher order chromatin structures. This gene is intronless and encodes a replication-dependent histone that is a member of the histone H3 family. Transcripts from this gene lack poly A tails; instead, they contain a palindromic termination element. This gene is located separately from the other H3 genes that are in the histone gene cluster on chromosome 6p22-p21.3. H3.4; H3/g; H3FT; H3t; HIST3H3; Histone H3; HIST1H3A ALTERNATE NAMES: ANTIBODY TYPE: Polyclonal HOST/ISOTYPE: Rabbit / IgG IMMUNOGEN: A synthetic methylated peptide targeting residues around Arginine 8 of human histone H3 MOLECULAR WEIGHT: 18 kDa PURIFICATION: Affinity purified FORM: Liquid FORMULATION: In PBS with 0.02% sodium azide, 50% glycerol, pH7.3 SPECIES REACTIVITY: Human, Mouse, Rat STORAGE CONDITIONS: Store at -20ºC. Avoid freeze / thaw cycles APPLICATIONS AND USAGE: WB 1:500 - 1:2000, IF 1:50 - 1:200 Note: This information is only intended as a guide. The optimal dilutions must be determined by the user Western blot analysis of H3R8me2(asym) expression in Dot-blot analysis of methylation peptides using Anti- HeLa cells and H3 protein. -

Theranostics CAPG Enhances Breast Cancer Metastasis by Competing
Theranostics 2018, Vol. 8, Issue 9 2549 Ivyspring International Publisher Theranostics 2018; 8(9): 2549-2564. doi: 10.7150/thno.22523 Research Paper CAPG enhances breast cancer metastasis by competing with PRMT5 to modulate STC-1 transcription Sheng Huang1,2#, Yayun Chi1,#, Yi Qin3,#, Ziliang Wang4, Bingqiu Xiu1, Yonghui Su1, Rong Guo1, Liang Guo1, Hefen Sun1, Chujia Zeng1, Shuling Zhou5, Xin Hu1, Sheng Liu6, Zhimin Shao1, Zhaohui Wu7, Wei Jin1,, Jiong Wu1, Department of Breast Surgery, Breast Cancer Institute, Shanghai Cancer Center, Department of Oncology, Shanghai Medical College, Fudan University, Shanghai, People's Republic of China The Second Department of Breast Surgery, The Third Affiliated Hospital of Kunming Medical University (Tumor Hospital of Yunnan Province), Kunming, Yunnan, People's Republic of China Department of Pancreas & Hepatobiliary Surgery, Pancreas & Hepatobiliary Cancer Institute, Shanghai Cancer Center, Department of Oncology, Shanghai Medical College, Fudan University, Shanghai, People's Republic of China Cancer Institute, Shanghai Cancer Center, Department of Oncology, Shanghai Medical College, Fudan University, Shanghai, People’s Republic of China Department of Pathology, Shanghai Cancer Center, Fudan University, Shanghai, People's Republic of China Department of Breast Surgery and Pharmacology Laboratory of Traditional Chinese Medicine, Long Hua Hospital, Shanghai University of Traditional Chinese Medicine, Shanghai, People's Republic of China Department of Pathology and Laboratory Medicine; Center for Cancer Research, University of Tennessee Health Science Center, Memphis, Tennessee # These authors contributed equally to this work. Corresponding author: Jiong Wu, Department of Breast Surgery, Building 2, No. 270 Dong An Road, Shanghai, 200032 Email: [email protected]; Tel: 86-21-64175590-3423; Fax: 86-21-64172585 or Wei Jin, Breast Cancer Institute, Building 7, No. -

Anti-H3r8me2(Sym) Antibody
FOR RESEARCH USE ONLY! 01/20 Anti-H3R8me2(sym) Antibody CATALOG NO.: A2046-100 (100 µl) BACKGROUND DESCRIPTION: Histones are basic nuclear proteins that are responsible for the nucleosome structure of the chromosomal fiber in eukaryotes. Nucleosomes consist of approximately 146 bp of DNA wrapped around a histone octamer composed of pairs of each of the four core histones (H2A, H2B, H3, and H4). The chromatin fiber is further compacted through the interaction of a linker histone, H1, with the DNA between the nucleosomes to form higher order chromatin structures. This gene is intronless and encodes a replication-dependent histone that is a member of the histone H3 family. Transcripts from this gene lack poly A tails; instead, they contain a palindromic termination element. This gene is located separately from the other H3 genes that are in the histone gene cluster on chromosome 6p22-p21.3. H3.4; H3/g; H3FT; H3t; HIST3H3; Histone H3; HIST1H3A ALTERNATE NAMES: ANTIBODY TYPE: Polyclonal HOST/ISOTYPE: Rabbit / IgG IMMUNOGEN: A synthetic methylated peptide targeting residues around Arginine 8 of human Histone H3 MOLECULAR WEIGHT: 18 kDa PURIFICATION: Affinity purified FORM: Liquid FORMULATION: In PBS with 0.02% sodium azide, 50% glycerol, pH 7.3 SPECIES REACTIVITY: Human, Mouse, Rat STORAGE CONDITIONS: Store at -20ºC. Avoid freeze / thaw cycles APPLICATIONS AND USAGE: WB 1:500 - 1:2000, IF 1:50 - 1:200 Note: This information is only intended as a guide. The optimal dilutions must be determined by the user Western blot analysis of H3R8me2(sym) expression in Dot-blot analysis of methylation peptides using Anti- HeLa cells and H3 protein.