Innsbruck Universities
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
Wiener Neustadt in MOTION!
Wiener Neustadt Discover. Experience. Live. CITY IN MOTION! ATTRACTIONS AND TOURS EVERYTHING IS NEW IN WIENER NEUSTADT The birthplace of Emperor Maximilian I, Wiener Neustadt is of great historical significance. The history of Wiener Neustadt, spanning centu- ries, is so exciting that it is quite surprising that the gates to the important sights were not opened much earlier. However, because of this, the Lower Austrian Provincial Exhibition 2019 sparked an CONTENTS initial, veritable rush to visit these, difficult to access, historical locations. Here you will find a complete overview of the Attractions ...........................................Page 4 sights and museums of Wiener Neustadt. Freely accessible highlights ................... Page 15 Culinary delights are not to be neglected either. Museums .............................................Page 18 Wiener Neustadt has an extraordinary variety of bars and restaurants to offer: from the rustic Getting there and parking ...................... Page 28 Stadtheurigen to the Beisl and Gasthaus with Useful contacts ....................................Page 32 everything to home-style cooking to top level cui- Town map .............................................Page 34 sine. Numerous coffee houses line the entire city center, inviting you to linger. 2 3 SIGHTSEEING THERESIAN MILITARY ACADEMY The castle was built about 50 years after the city was founded in 1192 as a military base for the last Babenberger, Friedrich II. Over the centuries the castle has been expanded and been given various new purposes. Emperor Friedrich III. had the castle fundamentally rebuilt, largely giving it its current appearance. Emperor Maximilian I was born and baptized in the castle in Wiener Neustadt and spent his youth here. From here the Holy Roman Empire was expanded. The empire grew so large that “the sun never set“. -

Schienenkorridore Für Die Steiermark Ausbauvorstellungen Für Den Güter- Und Personenverkehr
Steirische Regionalpolitische Studien Nr. 01/2018 Schienenkorridore für die Steiermark Ausbauvorstellungen für den Güter- und Personenverkehr Autoren: DI Dr.Helmut Adelsberger Dkfm. Dr. Heinz Petzmann Studie im Auftrag der steirischen Sozialpartner, März 2018 Inhaltsverzeichnis Seite Executive Summary 3 1. Das Transeuropäische Verkehrsnetz; Relevanz für die Steiermark 7 2. Raumstruktur und Lage der Steiermark im österreichischen und europäischen Schienennetz 14 2.1. Die Raumstruktur der Steiermark 14 2.2. Die Lage der Steiermark im österreichischen und europäischen Schienennetz 16 3. Bestehende Situation und Ziele 24 4. Der Baltisch-Adriatische Korridor 29 4.1. Bestand und aktuelle Planungen von der Ostsee bis einschließlich Wien 29 4.2. Bestand und aktuelle Planungen südlich von Wien bis Bruck an der Mur 31 4.3. Bestand und aktuelle Planungen südwestlich von Graz 36 4.4. Verkehrsströme im Baltisch-Adriatischen Korridor 39 4.5. Ausbaubedarf im Baltisch-Adriatischen Korridor 41 5. Die Pyhrnachse 45 5.1. Die angestrebte Verankerung der Pyhrnachse im TEN-T 45 5.2. Bestand und aktuelle Planungen nördlich von Bruck an der Mur 47 5.3. Bestand und aktuelle Planungen südlich von Werndorf 52 5.4. Verkehrsströme in der Pyhrnachse 56 5.5. Ausbaubedarf in der Pyhrnachse 59 5.6. Die Krapina-Bahn 62 6. Der Abschnitt Bruck an der Mur – Graz – Werndorf 68 6.1. Bestand und aktuelle Planungen 68 6.2. Verkehrsströme im Abschnitt Bruck an der Mur – Graz – Werndorf 73 6.3. Ausbaubedarf im Abschnitt Bruck an der Mur – Graz – Werndorf 74 7. Die Grazer Ostbahn 84 7.1. Bestand und aktuelle Planungen 84 7.2. Ausbaubedarf der Grazer Ostbahn 86 8. -

Stellungskundmachung 2020 NÖ
M I L I T Ä R K O M M A N D O N I E D E R Ö S T E R R E I C H Ergänzungsabteilung: 3101 St. PÖLTEN, Kommandogebäude Feldmarschall Hess, Schießstattring 8 Parteienverkehr: Mo-Do von 0800 bis 1400 Uhr und Fr von 0800 bis 1200 Uhr Telefon: 050201/30, Fax: 050201/30-17410 E-Mail: [email protected] STELLUNGSKUNDMACHUNG 2020 Auf Grund des § 18 Abs. 1 des Wehrgesetzes 2001 (WG 2001), BGBl. I Nr. 146, haben sich alle österreichischen Staatsbürger männlichen Geschlechtes des G E B U R T S J A H R G A N G E S 2 0 0 2 sowie alle älteren wehrpflichtigen Jahrgänge, die bisher der Stellungspflicht noch nicht nachgekommen sind, gemäß dem unten angeführten Plan der Stellung zu unterziehen. Österreichische Staatsbürger des Geburtsjahrganges 2002 oder eines älteren Geburtsjahrganges, bei denen die Stellungspflicht erst nach dem in dieser Stellungskundma- chung festgelegten Stellungstag entsteht, haben am 16.12.2020 zur Stellung zu erscheinen, sofern sie nicht vorher vom Militärkommando persönlich geladen wurden. Für Stellungspflichtige, welche ihren Hauptwohnsitz nicht in Österreich haben, gilt diese Stellungskundmachung nicht. Sie werden gegebenenfalls gesondert zur Stellung auf- gefordert. Für die Stellung ist insbesondere Folgendes zu beachten: 1. Für den Bereich des Militärkommandos NIEDERÖSTERREICH werden die Stellungspflichtigen durch die 3. Bei Vorliegen besonders schwerwiegender Gründe besteht die Möglichkeit, dass Stellungspflichtige auf ihren An- Stellungskommission NIEDERÖSTERREICH der Stellung unterzogen. Das Stellungsverfahren nimmt in der Regel trag in einem anderen Bundesland oder zu einem anderen Termin der Stellung unterzogen werden. 1 1/2 Tage in Anspruch. -

Fahrplan 2021 Fahrplanvorschau Bahn Und Bus Verbesserungen
Fahrplan 2021 Fahrplanvorschau Bahn und Bus Verbesserungen Stand November 2020 Alle Angaben ohne Gewähr! Inhalt 1. Wesentliche Neuerungen im Angebot 2. Änderungen im Detail 1. Westachse und Franz-Josefs-Bahn 2. Südachse 3. Ostachse und Nordburgenland 4. S-Bahn Wien und Nordäste 4 Wesentliche Neuerungen im Angebot Fahrplan 2021 gültig ab 13. Dezember 2020 Alle Angaben ohne Gewähr! Änderungen Fahrplan 2021: Überblick Laaer Ostbahn weitere Taktlückenschlüsse und täglicher Stundentakt Angebotsausweitungen im gesamten Netz • zusätzliche Abend-/Frühzüge auf diversen Strecken bis Laa/Thaya 4 Züge pro Stunde bis • zusätzliche HVZ-Verstärker u.a. auf Regionalbahnen St. Andrä-Wördern Weiterführung der S40-Verstärker Wien FJBf. – Kritzendorf als R40 bis St. Andrä-Wördern Nordbahn Stundentakt Mo – Fr bis Bernhardsthal bzw. Břeclav Tullnerfeldbahn: S40-Stundentakt auch am Wochenende St. Pölten – Traismauer – Tullnerfeld (weiter nach Wien FJBf.) Elektrifizierung Marchfeldbahn: Gänserndorf – Marchegg Weiterführung der S1 aus Wien von Gänserndorf nach Marchegg, täglicher Abendzüge auf der Rudolfsbahn Stundentakt Ausweitung des Stundentakts Amstetten – Waidhofen/Ybbs bis ~0:05 ab Amstetten Badner Bahn Wien Oper – Wiener Neudorf durchgehender 7,5-Minuten-Takt Neusiedler Seebahn Citybahn Waidhofen: Wien – Neusiedl/See - Pamhagen • Halbstundentakt (statt Stundentakt) • täglicher und durchgängiger • neuer Endpunkt Waidhofen Stundentakt Pestalozzistraße (Streckenkürzung) • Betriebszeitausweitung Innere Aspangbahn: Wien Hbf. – Traiskirchen – Wiener Neustadt -

Efpia Report
EFPIA REPORT Country: Austria Reporting Currency: EUR Reporting Year: 2016 Version: 03 Date Published: 17/07/2017 Fees for Service and Country Contribution to Costs of Events Donations Consultancy of Full Name City Principal Practice Address and Grants Total Principal Travel & to HCOs Registration Related Practice Sponsorship Accom- Fees Fees Expenses modation Health Care Professionals (HCPs) Agnes Loidl Innsbruck AT Innrain 43/Parterre 500 180 680 Agnes Silgoner Linz AT Seilerstätte 4 193 193 Agnieszka Ziomka Schwarzach im Pongau AT Kardinal Schwarzenberg-Straße 2-6 193 193 Ahmet Gökhan Uyanik Wien AT Heinrich Collin-Straße 30 758 800 1.558 Akós Heinemann Graz AT Heinrichstraße 31a 600 600 Alexander Fortelny Wien AT Eingang Löblichgasse 16 400 400 Alexander Gallee Vorderweißenbach AT Sonnenplatz 2 377 377 Alexander Stifter Hollabrunn AT Robert Löffler-Straße 20 193 193 Alexandra Pöll Hall in Tirol AT Milser Straße 10 400 400 Alexandra Weidinger Linz AT Karl Wiser-Straße 7/1 106 106 Alice Kafka Wien AT Auhofstraße 189 155 155 Ali Kaan Akmanlar Hallein AT Bürgermeisterstraße 34 174 174 Alois Günther Gastl Innsbruck AT Anichstraße 35 500 522 1.022 Alois Waschnig Leoben AT Schillerstraße 3 193 193 Amila Siljak Hallein AT Griesplatz 8 751 751 Andrea-Eva Bartok-Heinrich Wien AT Wienerbergstraße 13 300 300 Andrea Handke Graz AT Auenbruggerplatz 15 544 544 Andrea Mohn-Staudner Wien AT Sanatoriumstraße 2 500 500 Andrea Schadler Graz AT Elisabethinergasse 14 135 135 Andrea Scherr-Umundum Graz AT Elisabethinergasse 14 135 135 Andreas Kretschmer -

Erase Poster ÖKG 2021
Ertugliflozin to Reduce Arrhythmic burden in ICD/CRT patientS (ERASe-Trial) – a phase III Study Dirk von Lewinski1, Norbert J Tripolt2, Harald Sourij2, Abderrahim Oulhaj3, Hannes Alber4, Marianne Gwechenberger5, Martin Martinek6, Michael Nürnberg7, Franz Xaver Roithinger8, Clemens Steinwender9, Markus Stühlinger10, Ursula Rohrer1, Martin Manninger-Wünscher1, Daniel Scherr, MD1 on behalf of the ERASe study group 1Medical University of Graz, Department of Internal Medicine, Division of Cardiology, 8010 Graz, Austria. 2Medical University of Graz, Department of Internal Medicine, Division of Endocrinology and Diabetology, 8010 Graz, Austria. 3College of Medicine and Health Sciences, United Arab Emirates University, P.O.Box 17666, Al-Ain, United Arab Emirates. 4Klinikum Klagenfurt, Abteilung für Innere Medizin und Kardiologie 5Medical University of Vienna, Department of Cardiology, 1090 Vienna, Austria. 6Ordensklinikum Linz Elisabethinen, Innere Medizin 2 mit Kardiologie, Angiologie und Intensivmedizin 7Klinik Ottakring, Abteilung für Kardiologie, Wien 8Landesklinikum Wiener Neustadt, Abteilung für Innere Medizin, Kardiologie und Nephrologie, Wiener Neustadt 9Kepler University Hospital Linz, Department of Cardiology and Intensive Care Medicine, Linz, Austria. 10Medical University of Innsbruck, Univ. Clinic of Internal Medicine III / Cardiology and Angiology, 6020 Innsbruck, Austria Hypothesis Inclusion and exclusion criteria Inclusion criteria Exclusion criteria Ø Treatment with the SGLT-2 inhibitor 1) HFrEF or HFmrEF and ICD±CRT therapy > -

University of Applied Sciences Wiener Neustadt, Austria
University of Applied Sciences Wiener Neustadt, Austria Business • Engineering • Health Studies • Security www.fhwn.ac.at “Zauberberge” – Magic Mountains, reached in 30 minutes from Wiener Neustadt Exchange Semester in Austria Austria – experience, history, arts, sports and leisure CRITICAL DATES The University of Applied Sciences alpine world that runs right up to the Wiener Neustadt, Austria, provides an town outskirts. The magic mountains > Application deadline excellent cultural and learning experi- with their steep cliff faces, the meadows ence for students interested in studying and woods offer exciting discovery tours, Winter semester abroad in Europe. Located in the heart of enabling exchange students to fully enjoy EU countries: 15th June Europe, Austria is a country steeped in the area in all its stunning beauty – st history and tradition. The city of Wiener cycling or walking through it or taking Non EU & Overseas: 1 June Neustadt is only a 35-min train ride from part in our skiing event. the capital city Vienna, where students Spring semester can spend time exploring the many his- From Wiener Neustadt travellers can EU Countries: 15th Nov. torical sites that Austria has to offer. reach Vienna International Airport by car Non EU & Overseas: 1st Nov. in 40 minutes, Hungary in 20 minutes, Besides sight-seeing along the famous Budapest in 2.5 hours, the Czech Repub- Ringstraße or visiting the imperial lic in 80 minutes, Salzburg in 3 hours, > Academic year palaces, one can spend the day shop- Munich in 5 hours and Venice in 6 hours. ping, sipping coffee at an outdoor café or Wiener Neustadt is the perfect location Winter Semester: taking advantage of the endless theatrical for trips to important cities of the former Beginning of Sept. -

Pdf Document
Local travel information for your trip to the ESSL Research and Training Centre in Wiener Neustadt, Austria Airport and Trains The closest international airport is VIE (Vienna International Airport) with a lot of direct connections to destinations in Europe and worldwide. From the airport there are frequent train connections to Vienna, either by the City Airport Train (CAT) to central Vienna, local commuter trains (S-Bahn) or fast direct trains to Vienna Main Train Station (Wien Hauptbahnhof). In most cases you will prefer a train via Wien Hauptbahnhof, because from there you have direct fast connections to Wiener Neustadt (sometimes abbreviated with Wr. Neustadt). Please use the following website, the route planner and online ticket shop of the Austrian Railways OeBB, to find your best connection from the airport or from Vienna to Wiener Neustadt: http://fahrplan.oebb.at/bin/query.exe/en Enter from “Flughafen Wien” to “Wiener Neustadt Hauptbahnhof” and be sure to take a fast connection with only one change in Vienna. In case you miss one train, no problem, simply take the next one, your ticket is valid in all trains. Wiener Neustadt Although the mostly booked hotels are not far from the train station (Wiener Neustadt Hauptbahnhof), you can choose a taxi in Wiener Neustadt, if you prefer to bring your luggage easily to the hotel. The taxi stand is situated in front of the main exit of the train station. You can also order a taxi via phone: +43 2622 25500 or +43 2622 22000 Help, I am lost In case you need personal assistance in Austria, please call Katja Dorninger from the ESSL team. -

UPC TV MINI 18-08-2016 Wien Wiener Neustadt Baden
UPC TV MINI Entgeltbestimmungen und Leistungsbeschreibungen für Wien, Wiener Neustadt, Baden, Wien West, Oberösterreich, Graz und Klagenfurt Gültig ab 18.08.2016 Seite 1 von 6 Monatsentgelte UPC TV MINI UPC TV MINI 56 digitale TV Kanäle [davon 52 in SD (Standard Definition) & 4 in HD (High € 24,55 Definition)] ohne UPC MediaBox Für UPC TV Mini ist ein TV-Gerät mit DVB-C Tuner Voraussetzung. Hinweise: o UPC TV MINI ist nur mit 12 Monaten Mindestvertragsdauer erhältlich. o Das aktuelle Senderangebot entnehmen Sie bitte dem UPC TV MINI Folder und der Homepage www.upc.at o UPC ist berechtigt, dass Leistungsangebot jederzeit zu verändern, insbesondere TV-Programme und Radiosender auszutauschen oder zu entfernen, wobei zumindest 25 digital (SD) übermittelte TV-Programme als vereinbart gelten. o Bitte beachten Sie, dass bei ausländischen TV-Sendern die Programminhalte von den jeweiligen Ausstrahlungsrechten in Österreich abhängig sind, auf die UPC keinen Einfluss hat. Seite 2 von 6 Einmalige Entgelte Anschlussentgelt Für den Anschluss eines TV- bzw. Videogerätes. Das Anschlussentgelt beinhaltet die Einstellung der Programme an einem Fernsehgerät. Die Programmeinstellung zusätzlicher Fernseh- bzw. Videogeräte kostet € 14,00 pro Gerät. Die Standardverkabelung ist bei Anschluss gewährleistet. Sollten abweichende Kabellängen gewünscht werden, so können diese bei der Montage erworben werden. Die Kabelverbindung zwischen der UPC Steckdose und dem Fernseh- bzw. Radiogerät ist nicht im Anschlussentgelt enthalten. Anschlussentgelt € 69,90 Reduziertes Anschlussentgelt € 29,90 Aktivierungsentgelt Für jede Aktivierung/Freischaltung eines Produktes Aktivierungsentgelt € 34,90 Reduziertes Aktivierungsentgelt € 24,90 Die oben angeführten Anschluss- und Aktivierungsentgelte kommen in folgenden Fällen zur Verrechnung. Hinweis: Es werden höchstens 1 x Anschlussentgelt oder reduziertes Anschlussentgelt und 1 x Aktivierungsentgelt oder reduziertes Aktivierungsentgelt verrechnet. -
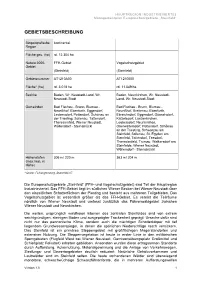
Gebietsbeschreibung
HAUPTREGION INDUSTRIEVIERTEL Managementplan Europaschutzgebiete „Steinfeld“ GEBIETSBESCHREIBUNG Biogeografische kontinental Region Fläche ges. (ha) rd. 12.304 ha Natura 2000- FFH-Gebiet Vogelschutzgebiet Gebiet (Steinfeld) (Steinfeld) Gebietsnummer AT1210A00 AT1210000 Fläche* (ha) rd. 3.018 ha rd. 11.549 ha Bezirke Baden, Wr. Neustadt-Land, Wr. Baden, Neunkirchen, Wr. Neustadt- Neustadt-Stadt Land, Wr. Neustadt-Stadt Gemeinden Bad Fischau - Brunn, Blumau - Bad Fischau - Brunn, Blumau - Neurißhof, Ebenfurth, Eggendorf, Neurißhof, Breitenau, Ebenfurth, Leobersdorf, Pottendorf, Schönau an Ebreichsdorf, Eggendorf, Günselsdorf, der Triesting, Sollenau, Tattendorf, Katzelsdorf, Lanzenkirchen, Theresienfeld, Wiener Neustadt, Leobersdorf, Neunkirchen, Wöllersdorf - Steinabrückl Oberwaltersdorf, Pottendorf, Schönau an der Triesting, Schwarzau am Steinfeld, Sollenau, St. Egyden am Steinfeld, Tattendorf, Teesdorf, Theresienfeld, Trumau, Weikersdorf am Steinfelde, Wiener Neustadt, Wöllersdorf - Steinabrückl Höhenstufen 306 m/ 220 m 363 m/ 204 m (max./min. m Höhe) * Quelle: Feinabgrenzung, Stand Mai 07 Die Europaschutzgebiete „Steinfeld” (FFH- und Vogelschutzgebiet) sind Teil der Hauptregion Industrieviertel. Das FFH-Gebiet liegt im südlichen Wiener Becken bei Wiener Neustadt über den eiszeitlichen Schotterfächern der Piesting und besteht aus mehreren Teilgebieten. Das Vogelschutzgebiet ist wesentlich größer als das FFH-Gebiet. Es vereint die Teilräume nördlich von Wiener Neustadt und umfasst zusätzlich das Föhrenwaldgebiet zwischen Wiener Neustadt -

Austria (1937 Borders)
Luftwaffe Airfields 1935-45 Luftwaffe Airfields 1935-45 Austria (1937 Borders) By Henry L. deZeng IV Graz Edition: June 2014 Luftwaffe Airfields 1935-45 Copyright © by Henry L. deZeng IV (Work in Progress). (1st Draft 2014) Blanket permission is granted by the author to researchers to extract information from this publication for their personal use in accordance with the generally accepted definition of fair use laws. Otherwise, the following applies: All rights reserved. No part of this publication, an original work by the authors, may be reproduced, stored in or introduced into a retrieval system, or transmitted, in any form, or by any means (electronic, mechanical, photocopying, recording or otherwise), without the prior written permission of the author. Any person who does any unauthorized act in relation to this publication may be liable to criminal prosecution and civil claims for damages. This information is provided on an "as is" basis without condition apart from making an acknowledgement of authorship. Luftwaffe Airfields 1935-45 Airfields Austria (1937 borders) Introduction Preface The Germans marched into Austria on 12 March 1938 and took possession of the country and its military by the end of the day. A report dated 25 March stated that the entire strength of the Austrian Air Force consisted of 243 aircraft, of which approximately 50% were unserviceable, and these were based at Aigen, Graz-Thalerhof, Klagenfurt-Annabichl, St. Pölten, Wels, Wien, Wien-Aspern, Wiener Neustadt and Zeltweg. Orders were issued by the Luftwaffe near the end of March to begin improving Austria’s existing airfields, nearly all of which were located in the eastern part of the country, and build at least 12 Einsatzhafen (operational airfields) and 4 Feldflugplätze (field airstrips). -

Getting to the Medaustron Site
Getting to the MedAustron Site Your Journey to MedAustron within Wiener Neustadt Below is a map showing the location of our MedAustron and the TFZ building in Wiener Neustadt and including other points of interest to help with orientation. Arrival by CAR: • Directions to MedAustron from Vienna (from north) BY CAR Take the A2 in direction Graz to the exit Wöllersdorf; at the roundabout take the second exit in direction Wiener Neustadt; at the next roundabout, take the second exit towards Wiener Neustadt (B21 /Gutensteiner-Bundesstraße); at the following roundabout, take the first exit (onto B17); and the next roundabout, take the third exit onto Nikolaus August Otto Straße; turn right at Prof. Dr. Stephan Koren Straße; turn left at Johannes Gutenberg Straße; then turn left and take the first exit at the roundabout; the MedAustron building will be on the right side. • Arrival to MedAustron from Graz (from south) BY CAR Take the A2 direction Wien to the junction Knoten Wiener Neustadt in the direction Eisenstadt/Wiener Neustadt/-Süd/Mattersburg; keep left, follow the signs to Wiener Neustadt/B17 and take the B17/Neunkirchner Straße; continue on the B1; ; turn right at Prof. Dr. Stephan Koren Straße; turn left at Johannes Gutenberg Straße; then turn left and take the first exit at the roundabout; the MedAustron building will be on the right side. Directions to MedAustron using Public Transportation (Train, Bus, Airplane) • Arrival to MedAustron BY TRAIN and BUS Train to Wiener Neustadt Main Train Station(Wr. Neustadt Hauptbahnhof) Please check the train schedule posted in the internet at www.oebb.at.