N6-Methyladenosine-Dependent Regulation of Messenger RNA Stability
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Caspases Switch Off M6a RNA Modification Pathway to Reactivate A
bioRxiv preprint doi: https://doi.org/10.1101/2020.11.12.377127; this version posted November 13, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. 6 1 Caspases switch off m A RNA modification pathway to reactivate a 2 ubiquitous human tumor virus 3 Kun Zhang1,2, Yucheng Zhang3, Jun Wan3,4,5 and Renfeng Li1,2,6,7,8* 4 1Philips Institute for Oral Health Research, School of Dentistry, Virginia Commonwealth 5 University, Richmond, Virginia, 23298, USA 6 2Department of Oral and Craniofacial Molecular Biology, School of Dentistry, Virginia 7 Commonwealth University, Richmond, Virginia, 23298, USA 8 3Department of Medical and Molecular Genetics, Indiana University School of Medicine, 9 Indianapolis, Indiana, 46202, USA 10 4Center for Computational Biology and Bioinformatics, Indiana University School of Medicine, 11 Indianapolis, Indiana, 46202, USA. 12 5Department of BioHealth Informatics, School of Informatics and Computing, Indiana University 13 – Purdue University at Indianapolis, Indianapolis, Indiana, 46202, USA 14 6Department of Microbiology and Immunology, School of Medicine, Virginia Commonwealth 15 University, Richmond, Virginia, 23298, USA 16 7Massey Cancer Center, Virginia Commonwealth University, Richmond, Virginia, 23298, USA. 17 8Lead Contact 18 19 *Corresponding author: [email protected] (RL) 20 1 bioRxiv preprint doi: https://doi.org/10.1101/2020.11.12.377127; this version posted November 13, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. -

Novel Candidate Genes of Thyroid Tumourigenesis Identified in Trk-T1 Transgenic Mice
Endocrine-Related Cancer (2012) 19 409–421 Novel candidate genes of thyroid tumourigenesis identified in Trk-T1 transgenic mice Katrin-Janine Heiliger*, Julia Hess*, Donata Vitagliano1, Paolo Salerno1, Herbert Braselmann, Giuliana Salvatore 2, Clara Ugolini 3, Isolde Summerer 4, Tatjana Bogdanova5, Kristian Unger 6, Gerry Thomas6, Massimo Santoro1 and Horst Zitzelsberger Research Unit of Radiation Cytogenetics, Helmholtz Zentrum Mu¨nchen, Ingolsta¨dter Landstr. 1, 85764 Neuherberg, Germany 1Istituto di Endocrinologia ed Oncologia Sperimentale del CNR, c/o Dipartimento di Biologia e Patologia Cellulare e Molecolare, Universita` Federico II, Naples 80131, Italy 2Dipartimento di Studi delle Istituzioni e dei Sistemi Territoriali, Universita` ‘Parthenope’, Naples 80133, Italy 3Division of Pathology, Department of Surgery, University of Pisa, 56100 Pisa, Italy 4Institute of Radiation Biology, Helmholtz Zentrum Mu¨nchen, 85764 Neuherberg, Germany 5Institute of Endocrinology and Metabolism, Academy of Medical Sciences of the Ukraine, 254114 Kiev, Ukraine 6Department of Surgery and Cancer, Imperial College London, Hammersmith Hospital, London W12 0HS, UK (Correspondence should be addressed to H Zitzelsberger; Email: [email protected]) *(K-J Heiliger and J Hess contributed equally to this work) Abstract For an identification of novel candidate genes in thyroid tumourigenesis, we have investigated gene copy number changes in a Trk-T1 transgenic mouse model of thyroid neoplasia. For this aim, 30 thyroid tumours from Trk-T1 transgenics were investigated by comparative genomic hybridisation. Recurrent gene copy number alterations were identified and genes located in the altered chromosomal regions were analysed by Gene Ontology term enrichment analysis in order to reveal gene functions potentially associated with thyroid tumourigenesis. In thyroid neoplasms from Trk-T1 mice, a recurrent gain on chromosomal bands 1C4–E2.3 (10.0% of cases), and losses on 3H1–H3 (13.3%), 4D2.3–E2 (43.3%) and 14E4–E5 (6.7%) were identified. -

YTHDF2 (P-13): Sc-162426
SAN TA C RUZ BI OTEC HNOL OG Y, INC . YTHDF2 (P-13): sc-162426 BACKGROUND SOURCE The YTH domain family protein family (YTHDF) includes YTHDF1, YTHDF2 YTHDF2 (P-13) is an affinity purified goat polyclonal antibody raised against and TYHDF3. YTHDF2 (YTH domain family, member 2), also designated high- a peptide mapping within an internal region of YTHDF2 of human origin. glucose-regulated protein 8, CLL-associated antigen KW-14 or renal carcino - ma antigen NY-REN-2, is a 579 amino acid protein that also contains one PRODUCT YTH domain and exists as two alternatively spliced isoforms. Expressed in Each vial contains 200 µg IgG in 1.0 ml of PBS with < 0.1% sodium azide pancreas, testis and placenta, YTHDF2 has been identified as a transloca - and 0.1% gelatin. tion partner gene for RUNX1 and is encoded by a gene mapping to human chromosome 1p35.3. Human chromosome 1 spans 260 million base pairs, Blocking peptide available for competition studies, sc-162426 P, (100 µg contains over 3,000 genes and comprises nearly 8% of the human genome. pep tide in 0.5 ml PBS containing < 0.1% sodium azide and 0.2% BSA). Chromosome 1 houses a large number of disease-associated genes, includ - ing those that are involved in familial adenomatous polyposis, Stickler syn - APPLICATIONS drome, Parkinson’s disease, Gaucher disease, schizophrenia and Usher syn - YTHDF2 (P-13) is recommended for detection of YTHDF2 of mouse, rat and drome. human origin by Western Blotting (starting dilution 1:200, dilution range 1:100-1:1000), immunoprecipitation [1-2 µg per 100-500 µg of total protein REFERENCES (1 ml of cell lysate)], immunofluorescence (starting dilution 1:50, dilution 1. -

The Neurodegenerative Diseases ALS and SMA Are Linked at The
Nucleic Acids Research, 2019 1 doi: 10.1093/nar/gky1093 The neurodegenerative diseases ALS and SMA are linked at the molecular level via the ASC-1 complex Downloaded from https://academic.oup.com/nar/advance-article-abstract/doi/10.1093/nar/gky1093/5162471 by [email protected] on 06 November 2018 Binkai Chi, Jeremy D. O’Connell, Alexander D. Iocolano, Jordan A. Coady, Yong Yu, Jaya Gangopadhyay, Steven P. Gygi and Robin Reed* Department of Cell Biology, Harvard Medical School, 240 Longwood Ave. Boston MA 02115, USA Received July 17, 2018; Revised October 16, 2018; Editorial Decision October 18, 2018; Accepted October 19, 2018 ABSTRACT Fused in Sarcoma (FUS) and TAR DNA Binding Protein (TARDBP) (9–13). FUS is one of the three members of Understanding the molecular pathways disrupted in the structurally related FET (FUS, EWSR1 and TAF15) motor neuron diseases is urgently needed. Here, we family of RNA/DNA binding proteins (14). In addition to employed CRISPR knockout (KO) to investigate the the RNA/DNA binding domains, the FET proteins also functions of four ALS-causative RNA/DNA binding contain low-complexity domains, and these domains are proteins (FUS, EWSR1, TAF15 and MATR3) within the thought to be involved in ALS pathogenesis (5,15). In light RNAP II/U1 snRNP machinery. We found that each of of the discovery that mutations in FUS are ALS-causative, these structurally related proteins has distinct roles several groups carried out studies to determine whether the with FUS KO resulting in loss of U1 snRNP and the other two members of the FET family, TATA-Box Bind- SMN complex, EWSR1 KO causing dissociation of ing Protein Associated Factor 15 (TAF15) and EWS RNA the tRNA ligase complex, and TAF15 KO resulting in Binding Protein 1 (EWSR1), have a role in ALS. -

80014 YTHDF2 Antibody
Revision 1 C 0 2 - t YTHDF2 Antibody a e r o t S Orders: 877-616-CELL (2355) [email protected] 4 Support: 877-678-TECH (8324) 1 0 Web: [email protected] 0 www.cellsignal.com 8 # 3 Trask Lane Danvers Massachusetts 01923 USA For Research Use Only. Not For Use In Diagnostic Procedures. Applications: Reactivity: Sensitivity: MW (kDa): Source: UniProt ID: Entrez-Gene Id: WB, IF-IC H M R Mk Endogenous 65 Rabbit Q9Y5A9 51441 Product Usage Information 3. Wang, X. et al. (2014) Nature 505, 117-20. 4. Shi, H. et al. (2017) Cell Res 27, 315-28. Application Dilution 5. Du, H. et al. (2016) Nat Commun 7, 12626. 6. Kennedy, E.M. et al. (2016) Cell Host Microbe 19, 675-85. Western Blotting 1:1000 Immunofluorescence (Immunocytochemistry) 1:50 - 1:100 Storage Supplied in 10 mM sodium HEPES (pH 7.5), 150 mM NaCl, 100 µg/ml BSA and 50% glycerol. Store at –20°C. Do not aliquot the antibody. Specificity / Sensitivity YTHDF2 Antibody recognizes endogenous levels of total YTHDF2 protein. Species Reactivity: Human, Mouse, Rat, Monkey Source / Purification Polyclonal antibodies are produced by immunizing animals with a synthetic peptide corresponding to residues surrounding Gly167 of human YTHDF2 protein. Antibodies are purified by peptide affinity chromatography. Background N6-methyladenosine (m6A) is an abundant RNA modification that plays an important role in mRNA splicing, processing, and stability. The m6A modification is specifically recognized by members of the YT521B homology (YTH) domain-containing family (YTHDF), consisting of YTHDF1, YTHDF2, and YTHDF3. All three members of the YTHDF family are primarily cytosolic proteins that share similar sequence and domain structure, including a conserved C-terminal YTH domain that specifically interacts with m6A (1). -

The RNA M a Reader YTHDF2 Controls NK Cell Anti-Tumor And
bioRxiv preprint doi: https://doi.org/10.1101/2021.04.26.441557; this version posted April 27, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. The RNA m6A reader YTHDF2 controls NK cell anti-tumor and anti-viral immunity Shoubao Ma1,2,10, Jiazhuo Yan1,3,10, Tasha Barr1, Jianying Zhang4, Zhenhua Chen5, Li-Shu Wang6, Joseph C. Sun7, Jianjun Chen5,8, Michael A Caligiuria,1,2,8, Jianhua Yua,1,2,8,9 Affiliations: 1Department of Hematology and Hematopoietic Cell Transplantation, City of Hope National Medical Center, Los Angeles, California 91010, USA. 2Hematologic Malignancies Research Institute, City of Hope National Medical Center, Los Angeles, California 91010, USA. 3Department of Gynecological Radiotherapy, Harbin Medical University Cancer Hospital, Harbin 150081, China. 4Department of Computational and Quantitative Medicine, City of Hope National Medical Center, Los Angeles, California 91010, USA. 5Department of Systems Biology, Beckman Research Institute, City of Hope, Los Angeles, California 91010, USA. 6Division of Hematology and Oncology, Department of Medicine, Medical College of Wisconsin, Milwaukee, Wisconsin 53226, USA. 7Immunology Program, Memorial Sloan Kettering Cancer Center, New York 10065, USA 8Comprehensive Cancer Center, City of Hope, Los Angeles, California 91010, USA. 9Department of Immuno-Oncology, Beckman Research Institute, City of Hope, Los Angeles, California 91010, USA. 10Contributed equally to this work. aTo whom correspondence should be addressed. Email: Jianhua Yu, PhD, [email protected]; Michael A Caligiuri, MD, [email protected] bioRxiv preprint doi: https://doi.org/10.1101/2021.04.26.441557; this version posted April 27, 2021. -

M6a Modification in RNA: Biogenesis, Functions and Roles in Gliomas
Zhang et al. Journal of Experimental & Clinical Cancer Research (2020) 39:192 https://doi.org/10.1186/s13046-020-01706-8 REVIEW Open Access m6A modification in RNA: biogenesis, functions and roles in gliomas Yuhao Zhang1†, Xiuchao Geng2†, Qiang Li3, Jianglong Xu1, Yanli Tan4, Menglin Xiao1, Jia Song5, Fulin Liu6*, Chuan Fang1* and Hong Wang1,2,7* Abstract The chemical modification of RNA is a newly discovered epigenetic regulation mechanism in cells and plays a crucial role in a variety of biological processes. N6-methyladenine (m6A) mRNA modification is the most abundant form of posttranscriptional RNA modification in eukaryotes. Through the development of m6A RNA sequencing, the relevant molecular mechanism of m6A modification has gradually been revealed. It has been found that the effect of m6A modification on RNA metabolism involves processing, nuclear export, translation and even decay. As the most common malignant tumour of the central nervous system, gliomas (especially glioblastoma) have a very poor prognosis, and treatment efficacy is not ideal even with the application of high-intensity treatment measures of surgery combined with chemoradiotherapy. Exploring the origin and development mechanisms of tumour cells from the perspective of tumour biogenesis has always been a hotspot in the field of glioma research. Emerging evidence suggests that m6A modification can play a key role in gliomas through a variety of mechanisms, providing more possibilities for early diagnosis and targeted therapy of gliomas. The aim of the present review is to focus on the research progress regarding the association between m6A modification and gliomas. And to provide a theoretical basis according to the currently available literature for further exploring this association. -

Interactome Analyses Revealed That the U1 Snrnp Machinery Overlaps
www.nature.com/scientificreports OPEN Interactome analyses revealed that the U1 snRNP machinery overlaps extensively with the RNAP II Received: 12 April 2018 Accepted: 24 May 2018 machinery and contains multiple Published: xx xx xxxx ALS/SMA-causative proteins Binkai Chi1, Jeremy D. O’Connell1,2, Tomohiro Yamazaki1, Jaya Gangopadhyay1, Steven P. Gygi1 & Robin Reed1 Mutations in multiple RNA/DNA binding proteins cause Amyotrophic Lateral Sclerosis (ALS). Included among these are the three members of the FET family (FUS, EWSR1 and TAF15) and the structurally similar MATR3. Here, we characterized the interactomes of these four proteins, revealing that they largely have unique interactors, but share in common an association with U1 snRNP. The latter observation led us to analyze the interactome of the U1 snRNP machinery. Surprisingly, this analysis revealed the interactome contains ~220 components, and of these, >200 are shared with the RNA polymerase II (RNAP II) machinery. Among the shared components are multiple ALS and Spinal muscular Atrophy (SMA)-causative proteins and numerous discrete complexes, including the SMN complex, transcription factor complexes, and RNA processing complexes. Together, our data indicate that the RNAP II/U1 snRNP machinery functions in a wide variety of molecular pathways, and these pathways are candidates for playing roles in ALS/SMA pathogenesis. Te neurodegenerative disease Amyotrophic Lateral Sclerosis (ALS) has no known treatment, and elucidation of disease mechanisms is urgently needed. Tis problem has been especially daunting, as mutations in greater than 30 genes are ALS-causative, and these genes function in numerous cellular pathways1. Tese include mitophagy, autophagy, cytoskeletal dynamics, vesicle transport, DNA damage repair, RNA dysfunction, apoptosis, and pro- tein aggregation2–6. -

Structural Basis for Selective Binding of M6a RNA by the YTHDC1 YTH Domain
Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain Chao Xu1#,*, Xiao Wang2,3#, Ke Liu1#, Ian A. Roundtree3,4, Wolfram Tempel1, Yanjun Li1, Zhike Lu2,3, Chuan He2,3*, Jinrong Min1,5* 1Structural Genomics Consortium, University of Toronto, 101 College Street, Toronto, ON, M5G 1L7, Canada. 2Department of Chemistry and Institute for Biophysical Dynamics, The University of Chicago, 929 East 57th Street, Chicago, Illinois 60637, USA. 3Howard Hughes Medical Institute, The University of Chicago, 929 East 57th Street, Chicago, Illinois 60637, USA. 4Medical Scientist Training Program and Department of Biochemistry and Molecular Biology, The University of Chicago, 929 East 57th Street, Chicago, Illinois 60637, USA. 5Department of Physiology, University of Toronto, Toronto, ON, Canada M5S 1A8. #: These authors contributed equally to the work. *: Corresponding author: [email protected], [email protected], [email protected] 1 Abstract N6-methyladenosine (m6A) is the most abundant internal modification of nearly all eukaryotic mRNAs, and has recently been reported to be recognized by the YTH domain containing proteins. Here we present the crystal structures of the YTH domain of YTHDC1 and its complex with an m6A-containing RNA. Our structural studies, together with PAR-CLIP and biochemical experiments reveal the specific mode of m6A-YTH binding, but also explain the preferential recognition of the GG(m6A)C sequences by YTHDC1. 2 Methylation of internal adenosines to form N6-methyladenosine (m6A) is a key processing event during maturation of eukaryotic messenger RNAs (mRNAs), in concert with 5’ capping, 3’ polyadenylation and splicing1. m6A is highly conserved across all eukaryotes from yeast2 to human3 within a G(m6A)C (70%) or A(m6A)C (30%) motif, and is physiologically essential to metazoans4-6. -

YTHDF2 Destabilizes M6a-Modified Neural-Specific Rnas to Restrain Differentiation in Induced Pluripotent Stem Cells Adam M
Downloaded from rnajournal.cshlp.org on October 3, 2021 - Published by Cold Spring Harbor Laboratory Press YTHDF2 destabilizes m6A-modified neural-specific RNAs to restrain differentiation in induced pluripotent stem cells Adam M. Heck1,2,‡, Joseph Russo2,†, Jeffrey Wilusz1,2, Erin Osborne Nishimura1,3 and Carol J. Wilusz1, 2,* 1Program in Cell & Molecular Biology, 2Department of Microbiology, Immunology & Pathology, 3Department of Biochemistry & Molecular Biology, Colorado State University, Fort Collins, CO 80523, USA * lead contact and corresponding author: [email protected]; Phone: 970-491-4919 ‡Present address: Fred Hutchinson Cancer Research Center, 1100 Fairview Ave, Seattle, WA 98109 †Present address: Pfizer Inc., Chesterfield, MO 63017, USA Abstract N6-methyladenosine (m6A) is an abundant post-transcriptional modification that can impact RNA fate via interactions with m6A-specific RNA binding proteins. Despite accumulating evidence that m6A plays an important role in modulating pluripotency, the influence of m6A reader proteins in pluripotency is less clear. Here, we report that YTHDF2, an m6A reader associated with mRNA degradation, is highly expressed in induced pluripotent stem cells (iPSCs) and down-regulated during neural differentiation. Through RNA sequencing, we identified a group of m6A-modified transcripts associated with neural development that are directly regulated by YTDHF2. Depletion of YTHDF2 in iPSCs leads to stabilization of these transcripts, loss of pluripotency and induction of neural-specific gene expression. Collectively, our results suggest YTHDF2 functions to restrain expression of neural-specific mRNAs in iPSCs and facilitate their rapid and coordinated upregulation during neural induction. These effects are both achieved by destabilization of the targeted transcripts. Introduction The ability of stem cells to maintain a state of self-renewal, yet also rapidly differentiate in response to external signals requires complex and coordinated control of global gene expression. -
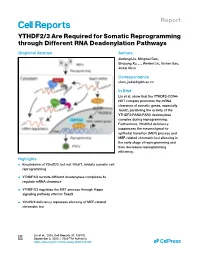
YTHDF2/3 Are Required for Somatic Reprogramming Through Different RNA Deadenylation Pathways
Report YTHDF2/3 Are Required for Somatic Reprogramming through Different RNA Deadenylation Pathways Graphical Abstract Authors Jiadong Liu, Mingwei Gao, Shuyang Xu, ..., Weiwei Liu, Xichen Bao, Jiekai Chen Correspondence [email protected] In Brief Liu et al. show that the YTHDF2-CCR4- NOT complex promotes the mRNA clearance of somatic genes, especially Tead2, paralleling the activity of the YTHDF3-PAN2-PAN3 deadenylase complex during reprogramming. Furthermore, Ythdf2/3 deficiency suppresses the mesenchymal-to- epithelial transition (MET) process and MEF-related chromatin loci silencing in the early stage of reprogramming and then decreases reprogramming efficiency. Highlights d Knockdown of Ythdf2/3, but not Ythdf1, inhibits somatic cell reprogramming d YTHDF2/3 recruits different deadenylase complexes to regulate mRNA clearance d YTHDF2/3 regulates the MET process through Hippo signaling pathway effector Tead2 d Ythdf2/3 deficiency represses silencing of MEF-related chromatin loci Liu et al., 2020, Cell Reports 32, 108120 September 8, 2020 ª 2020 The Author(s). https://doi.org/10.1016/j.celrep.2020.108120 ll ll OPEN ACCESS Report YTHDF2/3 Are Required for Somatic Reprogramming through Different RNA Deadenylation Pathways Jiadong Liu,1,2,3,7 Mingwei Gao,1,2,3,7 Shuyang Xu,1,2 Yaping Chen,1,2,3 Kaixin Wu,1,2,5 He Liu,1,2,4,5 Jie Wang,1,2 Xuejie Yang,4 Junwei Wang,1,2 Weiwei Liu,6 Xichen Bao,1,2,3,5 and Jiekai Chen1,2,3,4,5,8,* 1CAS Key Laboratory of Regenerative Biology, Guangzhou Institutes of Biomedicine and Health, Chinese Academy -

Anti-YTHDF2 Antibody (ARG40179)
Product datasheet [email protected] ARG40179 Package: 100 μl anti-YTHDF2 antibody Store at: -20°C Summary Product Description Rabbit Polyclonal antibody recognizes YTHDF2 Tested Reactivity Hu, Ms Tested Application WB Host Rabbit Clonality Polyclonal Isotype IgG Target Name YTHDF2 Antigen Species Human Immunogen Recombinant protein of Human YTHDF2. Conjugation Un-conjugated Alternate Names HGRG8; YTH domain-containing family protein 2; CLL-associated antigen KW-14; NY-REN-2; CAHL; Renal carcinoma antigen NY-REN-2; High-glucose-regulated protein 8 Application Instructions Application table Application Dilution WB 1:500 - 1:2000 Application Note * The dilutions indicate recommended starting dilutions and the optimal dilutions or concentrations should be determined by the scientist. Positive Control Mouse testis and Jurkat Calculated Mw 62 kDa Observed Size 68 kDa Properties Form Liquid Purification Affinity purified. Buffer PBS (pH 7.3), 0.02% Sodium azide and 50% Glycerol. Preservative 0.02% Sodium azide Stabilizer 50% Glycerol Storage instruction For continuous use, store undiluted antibody at 2-8°C for up to a week. For long-term storage, aliquot and store at -20°C. Storage in frost free freezers is not recommended. Avoid repeated freeze/thaw cycles. Suggest spin the vial prior to opening. The antibody solution should be gently mixed before use. Note For laboratory research only, not for drug, diagnostic or other use. www.arigobio.com 1/2 Bioinformation Gene Symbol YTHDF2 Gene Full Name YTH N(6)-methyladenosine RNA binding protein 2 Background This gene encodes a member of the YTH (YT521-B homology) superfamily containing YTH domain. The YTH domain is typical for the eukaryotes and is particularly abundant in plants.