Supporting Information
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Global Analysis Reveals the Complexity of the Human Glomerular Extracellular Matrix
Global analysis reveals the complexity of the human glomerular extracellular matrix Rachel Lennon,1,2 Adam Byron,1,* Jonathan D. Humphries,1 Michael J. Randles,1,2 Alex Carisey,1 Stephanie Murphy,1,2 David Knight,3 Paul E. Brenchley,2 Roy Zent,4,5 and Martin J. Humphries.1 1Wellcome Trust Centre for Cell-Matrix Research, Faculty of Life Sciences, University of Manchester, Manchester, UK; 2Faculty of Medical and Human Sciences, University of Manchester, Manchester, UK; 3Biological Mass Spectrometry Core Facility, Faculty of Life Sciences, University of Manchester, Manchester, UK; 4Division of Nephrology, Department of Medicine, Vanderbilt University Medical Center, Nashville, TN, USA; and 5Veterans Affairs Hospital, Nashville, TN, USA. *Present address: Edinburgh Cancer Research UK Centre, Institute of Genetics and Molecular Medicine, University of Edinburgh, Edinburgh, UK. Running title: Proteome of the glomerular matrix Word count: Abstract: 208, main text 2765 Corresponding author: Dr Rachel Lennon, Wellcome Trust Centre for Cell-Matrix Research, Michael Smith Building, University of Manchester, Manchester M13 9PT, UK. Phone: 0044 (0) 161 2755498. Fax: 0044 (0) 161 2755082. Email: [email protected] Abstract The glomerulus contains unique cellular and extracellular matrix (ECM) components, which are required for intact barrier function. Studies of the cellular components have helped to build understanding of glomerular disease; however, the full composition and regulation of glomerular ECM remains poorly understood. Here, we employed mass spectrometry–based proteomics of enriched ECM extracts for a global analysis of human glomerular ECM in vivo and identified a tissue-specific proteome of 144 structural and regulatory ECM proteins. This catalogue includes all previously identified glomerular components, plus many new and abundant components. -

Environmental Influences on Endothelial Gene Expression
ENDOTHELIAL CELL GENE EXPRESSION John Matthew Jeff Herbert Supervisors: Prof. Roy Bicknell and Dr. Victoria Heath PhD thesis University of Birmingham August 2012 University of Birmingham Research Archive e-theses repository This unpublished thesis/dissertation is copyright of the author and/or third parties. The intellectual property rights of the author or third parties in respect of this work are as defined by The Copyright Designs and Patents Act 1988 or as modified by any successor legislation. Any use made of information contained in this thesis/dissertation must be in accordance with that legislation and must be properly acknowledged. Further distribution or reproduction in any format is prohibited without the permission of the copyright holder. ABSTRACT Tumour angiogenesis is a vital process in the pathology of tumour development and metastasis. Targeting markers of tumour endothelium provide a means of targeted destruction of a tumours oxygen and nutrient supply via destruction of tumour vasculature, which in turn ultimately leads to beneficial consequences to patients. Although current anti -angiogenic and vascular targeting strategies help patients, more potently in combination with chemo therapy, there is still a need for more tumour endothelial marker discoveries as current treatments have cardiovascular and other side effects. For the first time, the analyses of in-vivo biotinylation of an embryonic system is performed to obtain putative vascular targets. Also for the first time, deep sequencing is applied to freshly isolated tumour and normal endothelial cells from lung, colon and bladder tissues for the identification of pan-vascular-targets. Integration of the proteomic, deep sequencing, public cDNA libraries and microarrays, delivers 5,892 putative vascular targets to the science community. -

Original Article FREM2 Is an Independent Predictor of Poor Survival in Clear Cell Renal Cell Carcinoma-Evidence from the Cancer Genome Atlas (TCGA)
Int J Clin Exp Med 2019;12(12):13741-13748 www.ijcem.com /ISSN:1940-5901/IJCEM0076963 Original Article FREM2 is an independent predictor of poor survival in clear cell renal cell carcinoma-evidence from the cancer genome atlas (TCGA) Weiping Huang, Yongyong Lu, Xixi Huang, Feng Wang, Zhixian Yu Department of Urology, The First Affiliated Hospital of Wenzhou Medical University, Wenzhou 325035, Zhejiang Province, China Received February 26, 2018; Accepted October 7, 2018; Epub December 15, 2019; Published December 30, 2019 Abstract: Fraser syndrome protein 1 (FRAS1) and FRAS1 related extracellular matrix protein 1 and 2 (FREM1, FREM2) are a novel group of basement membrane proteins. The relationship between the three gene (FRAS1, FREM1, FREM2) and renal clear cell carcinoma is completely unclear. Thus, in this research, we used the mRNA sequencing data derived from TCGA kidney renal clear cell carcinoma cohort to assess the association of FRAS1, FREM1 and FREM2 with different clinical features. FRAS1, FREM1 and FREM2 mRNA levels were downregulated in KIRC (kidney renal clear cell carcinoma) tissues than normal tissues (FRAS1, P < 0.0001; FREM1, P < 0.0001, FREM2, P = 0.0001), respectively. FRAS1, FREM1 and FREM2 were significantly different in histologic grade, patho- logic stage and pathologic T (all P < 0.001). Low FRAS1, FREM1 and FREM2 expression were correlated to worsen overall survival (all P < 0.01), and Low FREM1 and FREM2 expression had worse relapse-free survival (FREM1, P = 0.0113; FREM2, P = 0.0424). Multivariate Cox regression analysis revealed that FREM2 was an independent prog- nostic factor for overall survival. Taken together, FREM2 expression is an independent predictor of poor survival in renal clear cell carcinoma and is positively associated with advanced stage, high histologic grade. -

Supplemental Material Placed on This Supplemental Material Which Has Been Supplied by the Author(S) J Med Genet
BMJ Publishing Group Limited (BMJ) disclaims all liability and responsibility arising from any reliance Supplemental material placed on this supplemental material which has been supplied by the author(s) J Med Genet Supplement Supplementary Table S1: GENE MEAN GENE NAME OMIM SYMBOL COVERAGE CAKUT CAKUT ADTKD ADTKD aHUS/TMA aHUS/TMA TUBULOPATHIES TUBULOPATHIES Glomerulopathies Glomerulopathies Polycystic kidneys / Ciliopathies Ciliopathies / kidneys Polycystic METABOLIC DISORDERS AND OTHERS OTHERS AND DISORDERS METABOLIC x x ACE angiotensin-I converting enzyme 106180 139 x ACTN4 actinin-4 604638 119 x ADAMTS13 von Willebrand cleaving protease 604134 154 x ADCY10 adenylate cyclase 10 605205 81 x x AGT angiotensinogen 106150 157 x x AGTR1 angiotensin II receptor, type 1 106165 131 x AGXT alanine-glyoxylate aminotransferase 604285 173 x AHI1 Abelson helper integration site 1 608894 100 x ALG13 asparagine-linked glycosylation 13 300776 232 x x ALG9 alpha-1,2-mannosyltransferase 606941 165 centrosome and basal body associated x ALMS1 606844 132 protein 1 x x APOA1 apolipoprotein A-1 107680 55 x APOE lipoprotein glomerulopathy 107741 77 x APOL1 apolipoprotein L-1 603743 98 x x APRT adenine phosphoribosyltransferase 102600 165 x ARHGAP24 Rho GTPase-Activation protein 24 610586 215 x ARL13B ADP-ribosylation factor-like 13B 608922 195 x x ARL6 ADP-ribosylation factor-like 6 608845 215 ATPase, H+ transporting, lysosomal V0, x ATP6V0A4 605239 90 subunit a4 ATPase, H+ transporting, lysosomal x x ATP6V1B1 192132 163 56/58, V1, subunit B1 x ATXN10 ataxin -

Chemical Agent and Antibodies B-Raf Inhibitor RAF265
Supplemental Materials and Methods: Chemical agent and antibodies B-Raf inhibitor RAF265 [5-(2-(5-(trifluromethyl)-1H-imidazol-2-yl)pyridin-4-yloxy)-N-(4-trifluoromethyl)phenyl-1-methyl-1H-benzp{D, }imidazol-2- amine] was kindly provided by Novartis Pharma AG and dissolved in solvent ethanol:propylene glycol:2.5% tween-80 (percentage 6:23:71) for oral delivery to mice by gavage. Antibodies to phospho-ERK1/2 Thr202/Tyr204(4370), phosphoMEK1/2(2338 and 9121)), phospho-cyclin D1(3300), cyclin D1 (2978), PLK1 (4513) BIM (2933), BAX (2772), BCL2 (2876) were from Cell Signaling Technology. Additional antibodies for phospho-ERK1,2 detection for western blot were from Promega (V803A), and Santa Cruz (E-Y, SC7383). Total ERK antibody for western blot analysis was K-23 from Santa Cruz (SC-94). Ki67 antibody (ab833) was from ABCAM, Mcl1 antibody (559027) was from BD Biosciences, Factor VIII antibody was from Dako (A082), CD31 antibody was from Dianova, (DIA310), and Cot antibody was from Santa Cruz Biotechnology (sc-373677). For the cyclin D1 second antibody staining was with an Alexa Fluor 568 donkey anti-rabbit IgG (Invitrogen, A10042) (1:200 dilution). The pMEK1 fluorescence was developed using the Alexa Fluor 488 chicken anti-rabbit IgG second antibody (1:200 dilution).TUNEL staining kits were from Promega (G2350). Mouse Implant Studies: Biopsy tissues were delivered to research laboratory in ice-cold Dulbecco's Modified Eagle Medium (DMEM) buffer solution. As the tissue mass available from each biopsy was limited, we first passaged the biopsy tissue in Balb/c nu/Foxn1 athymic nude mice (6-8 weeks of age and weighing 22-25g, purchased from Harlan Sprague Dawley, USA) to increase the volume of tumor for further implantation. -

Human Induced Pluripotent Stem Cell–Derived Podocytes Mature Into Vascularized Glomeruli Upon Experimental Transplantation
BASIC RESEARCH www.jasn.org Human Induced Pluripotent Stem Cell–Derived Podocytes Mature into Vascularized Glomeruli upon Experimental Transplantation † Sazia Sharmin,* Atsuhiro Taguchi,* Yusuke Kaku,* Yasuhiro Yoshimura,* Tomoko Ohmori,* ‡ † ‡ Tetsushi Sakuma, Masashi Mukoyama, Takashi Yamamoto, Hidetake Kurihara,§ and | Ryuichi Nishinakamura* *Department of Kidney Development, Institute of Molecular Embryology and Genetics, and †Department of Nephrology, Faculty of Life Sciences, Kumamoto University, Kumamoto, Japan; ‡Department of Mathematical and Life Sciences, Graduate School of Science, Hiroshima University, Hiroshima, Japan; §Division of Anatomy, Juntendo University School of Medicine, Tokyo, Japan; and |Japan Science and Technology Agency, CREST, Kumamoto, Japan ABSTRACT Glomerular podocytes express proteins, such as nephrin, that constitute the slit diaphragm, thereby contributing to the filtration process in the kidney. Glomerular development has been analyzed mainly in mice, whereas analysis of human kidney development has been minimal because of limited access to embryonic kidneys. We previously reported the induction of three-dimensional primordial glomeruli from human induced pluripotent stem (iPS) cells. Here, using transcription activator–like effector nuclease-mediated homologous recombination, we generated human iPS cell lines that express green fluorescent protein (GFP) in the NPHS1 locus, which encodes nephrin, and we show that GFP expression facilitated accurate visualization of nephrin-positive podocyte formation in -

Nephronectin As a Matrix Effector in Cancer
cancers Review Nephronectin as a Matrix Effector in Cancer Synnøve Norvoll Magnussen 1,* , Jimita Toraskar 2,3 , Elin Hadler-Olsen 1,4, Tonje S. Steigedal 2,3 and Gunbjørg Svineng 1 1 Department of Medical Biology, Faculty of Health Sciences, UiT—The Arctic University of Norway, 9037 Tromsø, Norway; [email protected] (E.H.-O.); [email protected] (G.S.) 2 Department of Clinical and Molecular Medicine, Faculty of Medicine and Health Sciences, Norwegian University of Science and Technology (NTNU), 7491 Trondheim, Norway; [email protected] (J.T.); [email protected] (T.S.S.) 3 Cancer Clinic, St. Olavs Hospital HF, 7006 Trondheim, Norway 4 The Public Dental Health Service Competence Center of Northern Norway, 9271 Tromsø, Norway * Correspondence: [email protected] Simple Summary: The extracellular matrix provides an important scaffold for cells and tissues of multicellular organisms. The scaffold not only provides a secure anchorage point, but also functions as a reservoir for signalling molecules, sequestered and released when necessary. A dysregulated extracellular matrix may therefore modulate cellular behaviour, as seen during cancer progression. The extracellular matrix protein nephronectin was discovered two decades ago and found to regulate important embryonic developmental processes. Loss of either nephronectin or its receptor, integrin α8β1, leads to underdeveloped kidneys. Recent findings show that nephronectin is also dysregulated in breast cancer and plays a role in promoting metastasis. To enable therapeutic intervention, it is important to fully understand the role of nephronectin and its receptors in cancer progression. In Citation: Magnussen, S.N.; this review, we summarise the literature on nephronectin, analyse the structure and domain-related Toraskar, J.; Hadler-Olsen, E.; functions of nephronectin and link these functions to potential roles in cancer progression. -
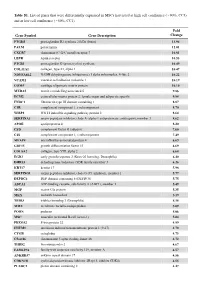
Table S1. List of Genes That Were Differentially Expressed in Mscs Harvested at High Cell Confluence (~90%, CC3) and at Low Cell Confluence (~50%, CC1)
Table S1. List of genes that were differentially expressed in MSCs harvested at high cell confluence (~90%, CC3) and at low cell confluence (~50%, CC1). Fold Gene Symbol Gene Description Change PTGDS prostaglandin D2 synthase 21kDa (brain) 13.94 PALM paralemmin 12.01 CXCR7 chemokine (C-X-C motif) receptor 7 10.81 LEPR leptin receptor 10.53 PTGIS prostaglandin I2 (prostacyclin) synthase 10.49 COL11A1 collagen, type XI, alpha 1 10.47 NDUFA4L2 NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4-like 2 10.22 VCAM1 vascular cell adhesion molecule 1 10.19 COMP cartilage oligomeric matrix protein 10.10 MXRA5 matrix-remodelling associated 5 9.86 ECM2 extracellular matrix protein 2, female organ and adipocyte specific 9.84 FNDC1 fibronectin type III domain containing 1 8.87 C1R complement component 1, r subcomponent 8.70 WISP2 WNT1 inducible signaling pathway protein 2 8.64 SERPINA3 serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3 8.62 APOE apolipoprotein E 8.20 CFD complement factor D (adipsin) 7.60 C1S complement component 1, s subcomponent 7.49 MFAP4 microfibrillar-associated protein 4 6.69 GDF15 growth differentiation factor 15 6.69 COL8A2 collagen, type VIII, alpha 2 6.68 EGR2 early growth response 2 (Krox-20 homolog, Drosophila) 6.40 DHRS3 dehydrogenase/reductase (SDR family) member 3 6.26 KRT17 keratin 17 5.98 SERPING1 serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 5.77 DEPDC6 DEP domain containing 6 (DEPDC6) 5.75 ABCA1 ATP-binding cassette, sub-family A (ABC1), member 1 5.49 MGP matrix -

Fibroblasts from the Human Skin Dermo-Hypodermal Junction Are
cells Article Fibroblasts from the Human Skin Dermo-Hypodermal Junction are Distinct from Dermal Papillary and Reticular Fibroblasts and from Mesenchymal Stem Cells and Exhibit a Specific Molecular Profile Related to Extracellular Matrix Organization and Modeling Valérie Haydont 1,*, Véronique Neiveyans 1, Philippe Perez 1, Élodie Busson 2, 2 1, 3,4,5,6, , Jean-Jacques Lataillade , Daniel Asselineau y and Nicolas O. Fortunel y * 1 Advanced Research, L’Oréal Research and Innovation, 93600 Aulnay-sous-Bois, France; [email protected] (V.N.); [email protected] (P.P.); [email protected] (D.A.) 2 Department of Medical and Surgical Assistance to the Armed Forces, French Forces Biomedical Research Institute (IRBA), 91223 CEDEX Brétigny sur Orge, France; [email protected] (É.B.); [email protected] (J.-J.L.) 3 Laboratoire de Génomique et Radiobiologie de la Kératinopoïèse, Institut de Biologie François Jacob, CEA/DRF/IRCM, 91000 Evry, France 4 INSERM U967, 92260 Fontenay-aux-Roses, France 5 Université Paris-Diderot, 75013 Paris 7, France 6 Université Paris-Saclay, 78140 Paris 11, France * Correspondence: [email protected] (V.H.); [email protected] (N.O.F.); Tel.: +33-1-48-68-96-00 (V.H.); +33-1-60-87-34-92 or +33-1-60-87-34-98 (N.O.F.) These authors contributed equally to the work. y Received: 15 December 2019; Accepted: 24 January 2020; Published: 5 February 2020 Abstract: Human skin dermis contains fibroblast subpopulations in which characterization is crucial due to their roles in extracellular matrix (ECM) biology. -

Architectural Delineation and Molecular Identification Of
© 2017. Published by The Company of Biologists Ltd | Biology Open (2017) 6, 1383-1390 doi:10.1242/bio.026336 RESEARCH ARTICLE Architectural delineation and molecular identification of extracellular matrix in ascidian embryos and larvae Jiankai Wei1, Guilin Wang1, Xiang Li1, Ping Ren1, Haiyan Yu1 and Bo Dong1,2,3,* ABSTRACT date, 28 types of collagens and >40 distinct α-chains have been The extracellular matrix (ECM) not only provides essential physical identified in vertebrates (Ricard-Blum, 2011). Collagens play scaffolding for cellular constituents but also initiates crucial structural roles and contribute to mechanical properties, biochemical and biomechanical cues that are required for tissue organization and pattern shaping of tissues. Proteoglycans are morphogenesis. In this study, we utilized wheat germ agglutinin characterized by a core protein that is covalently linked to (WGA) staining to characterize the ECM architecture in ascidian glycosaminoglycans (GAGs), which are long, negatively charged embryos and larvae. The results showed three distinct populations of and linear chains of disaccharide repeats. The primary biological ECM presenting in Ciona embryogenesis: the outer layer localized at function of proteoglycans derives from the biochemical and the surface of embryo, an inner layer of notochord sheath and the hydrodynamic characteristics of the GAGs, which bind water to apical ECM secreted by the notochord. To further elucidate the provide hydration and compressive resistance (Mouw et al., 2014). precise structure of Ciona -

Perkinelmer Genomics to Request the Saliva Swab Collection Kit for Patients That Cannot Provide a Blood Sample As Whole Blood Is the Preferred Sample
Autism and Intellectual Disability TRIO Panel Test Code TR002 Test Summary This test analyzes 2429 genes that have been associated with Autism and Intellectual Disability and/or disorders associated with Autism and Intellectual Disability with the analysis being performed as a TRIO Turn-Around-Time (TAT)* 3 - 5 weeks Acceptable Sample Types Whole Blood (EDTA) (Preferred sample type) DNA, Isolated Dried Blood Spots Saliva Acceptable Billing Types Self (patient) Payment Institutional Billing Commercial Insurance Indications for Testing Comprehensive test for patients with intellectual disability or global developmental delays (Moeschler et al 2014 PMID: 25157020). Comprehensive test for individuals with multiple congenital anomalies (Miller et al. 2010 PMID 20466091). Patients with autism/autism spectrum disorders (ASDs). Suspected autosomal recessive condition due to close familial relations Previously negative karyotyping and/or chromosomal microarray results. Test Description This panel analyzes 2429 genes that have been associated with Autism and ID and/or disorders associated with Autism and ID. Both sequencing and deletion/duplication (CNV) analysis will be performed on the coding regions of all genes included (unless otherwise marked). All analysis is performed utilizing Next Generation Sequencing (NGS) technology. CNV analysis is designed to detect the majority of deletions and duplications of three exons or greater in size. Smaller CNV events may also be detected and reported, but additional follow-up testing is recommended if a smaller CNV is suspected. All variants are classified according to ACMG guidelines. Condition Description Autism Spectrum Disorder (ASD) refers to a group of developmental disabilities that are typically associated with challenges of varying severity in the areas of social interaction, communication, and repetitive/restricted behaviors. -

Transcriptional Profiles of Differentiating Periocular Neural
ABSTRACT Transcriptional profiles of differentiating periocular neural crest cells and the function of Nephronectin during chick corneal development by Lian Bi During eye formation, periocular neural crest cells (pNC) migrate and differentiate to form the anterior ocular structures. In the chick cornea, this process involves two waves of migration that result in the formation of the corneal endothelium and stroma. Abnormalities in pNC migration lead to corneal malformation, such as anterior segment dystrophy. Corneal dystrophies, infections, and injuries can lead to corneal blindness, one of the major causes of blindness. Alternative treatments are developed because of the limitation of traditional corneal transplantation. These treatments benefit from the study of the molecular basis of corneal development and regeneration. However, corneal development is not fully understood. The purpose of this work was to elucidate the gene expression profiles during pNC migration and to examine the function of a highly regulated gene, Nephronectin (NPNT), during corneal formation. By performing RNA-seq analysis comparing pNC to the derived corneal structures, I analyzed differentially expressed genes and examined differentiated pathways during corneal formation. This project was designed to summarize the transcriptional regulation that happens at three levels: signaling pathways, transcription factors, and the downstream endothelial and stromal genes, providing gene candidates involved in corneal formation for future studies. From the RNA-seq analysis, I identified novel upregulation of NPNT among the extracellular matrix (ECM) proteins of the cornea. NPNT has been studied in other developmental processes but has not been linked to the corneal formation. I report that NPNT is distributed in the primary stroma during pNC migration.