Molecular Virology Module Specification 2020-21
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Molecular Biology for Computer Scientists
CHAPTER 1 Molecular Biology for Computer Scientists Lawrence Hunter “Computers are to biology what mathematics is to physics.” — Harold Morowitz One of the major challenges for computer scientists who wish to work in the domain of molecular biology is becoming conversant with the daunting intri- cacies of existing biological knowledge and its extensive technical vocabu- lary. Questions about the origin, function, and structure of living systems have been pursued by nearly all cultures throughout history, and the work of the last two generations has been particularly fruitful. The knowledge of liv- ing systems resulting from this research is far too detailed and complex for any one human to comprehend. An entire scientific career can be based in the study of a single biomolecule. Nevertheless, in the following pages, I attempt to provide enough background for a computer scientist to understand much of the biology discussed in this book. This chapter provides the briefest of overviews; I can only begin to convey the depth, variety, complexity and stunning beauty of the universe of living things. Much of what follows is not about molecular biology per se. In order to 2ARTIFICIAL INTELLIGENCE & MOLECULAR BIOLOGY explain what the molecules are doing, it is often necessary to use concepts involving, for example, cells, embryological development, or evolution. Bi- ology is frustratingly holistic. Events at one level can effect and be affected by events at very different levels of scale or time. Digesting a survey of the basic background material is a prerequisite for understanding the significance of the molecular biology that is described elsewhere in the book. -

Department of Pharmacology (GRAD) 1
Department of Pharmacology (GRAD) 1 faculty participate fully at all levels. The department has the highest DEPARTMENT OF level of NIH funding of all pharmacology departments and a great diversity of research areas is available to trainees. These areas PHARMACOLOGY (GRAD) include cell surface receptors, G proteins, protein kinases, and signal transduction mechanisms; neuropharmacology; nucleic acids, cancer, Contact Information and antimicrobial pharmacology; and experimental therapeutics. Cell and molecular approaches are particularly strong, but systems-level research Department of Pharmacology such as behavioral pharmacology and analysis of knock-in and knock-out Visit Program Website (http://www.med.unc.edu/pharm/) mice is also well-represented. Excellent physical facilities are available for all research areas. Henrik Dohlman, Chair Students completing the training program will have acquired basic The Department of Pharmacology offers a program of study that leads knowledge of pharmacology and related fields, in-depth knowledge in to the degree of doctor of philosophy in pharmacology. The curriculum is their dissertation research area, the ability to evaluate scientific literature, individualized in recognition of the diverse backgrounds and interests of mastery of a variety of laboratory procedures, skill in planning and students and the broad scope of the discipline of pharmacology. executing an important research project in pharmacology, and the ability The department offers a variety of research areas including to communicate results, analysis, and interpretation. These skills provide a sound basis for successful scientific careers in academia, government, 1. Receptors and signal transduction or industry. 2. Ion channels To apply to BBSP, students must use The Graduate School's online 3. -

Molecular Biology and Applied Genetics
MOLECULAR BIOLOGY AND APPLIED GENETICS FOR Medical Laboratory Technology Students Upgraded Lecture Note Series Mohammed Awole Adem Jimma University MOLECULAR BIOLOGY AND APPLIED GENETICS For Medical Laboratory Technician Students Lecture Note Series Mohammed Awole Adem Upgraded - 2006 In collaboration with The Carter Center (EPHTI) and The Federal Democratic Republic of Ethiopia Ministry of Education and Ministry of Health Jimma University PREFACE The problem faced today in the learning and teaching of Applied Genetics and Molecular Biology for laboratory technologists in universities, colleges andhealth institutions primarily from the unavailability of textbooks that focus on the needs of Ethiopian students. This lecture note has been prepared with the primary aim of alleviating the problems encountered in the teaching of Medical Applied Genetics and Molecular Biology course and in minimizing discrepancies prevailing among the different teaching and training health institutions. It can also be used in teaching any introductory course on medical Applied Genetics and Molecular Biology and as a reference material. This lecture note is specifically designed for medical laboratory technologists, and includes only those areas of molecular cell biology and Applied Genetics relevant to degree-level understanding of modern laboratory technology. Since genetics is prerequisite course to molecular biology, the lecture note starts with Genetics i followed by Molecular Biology. It provides students with molecular background to enable them to understand and critically analyze recent advances in laboratory sciences. Finally, it contains a glossary, which summarizes important terminologies used in the text. Each chapter begins by specific learning objectives and at the end of each chapter review questions are also included. -

Independent Research Resources Demonstrations/Simulations
Independent Research Resources Independent Generation of Research (IGoR) - IGoR provides a platform for people to pool their knowledge, resources, time, and creativity so that everyone can pursue their own scientific curiosity. Virginia Junior Academy of Science Resource Library - Extensive collection of open-access resources for students in Biology & Medicine, Botany, Ecology, Environmental Sciences, Chemistry, Engineering and Physics The Society for Science and the Public Science Project Resources - A catalog of science resources that can support your quest to learn and do science Science Buddies - Ideas for science projects Teacher resources National Center for Science Education Scientist in the Classroom - Platform allows teachers to request classroom visits from scientists Genetics Education Outreach Network (GEON) - Network of genetics professionals HHMI BioInteractive Data Points - Explore and interpret primary data from published research Biotech in a Box Loan Kits - Shipped to your school from Fralin Life Sciences Institute at Virginia Tech Demonstrations/Simulations Genetic Science Learning Center- Simulations, videos and interactive activities that explore genetics, cell biology, neuroscience, ecology and health Remotely Accessible Instruments for Nanotechnology (RAIN) - Access and control nanoinstruments over the Internet in real-time with the assistance of an experienced engineer PhET Simulations - Interactive STEM simulations for all grade levels HHMI BioInteractive Interactive Media - Recommendations: Virus Explorer; Exploring -

Biochemistry and Molecular Biology
School of Life Sciences / Department of Biochemistry and Biophysics BIOCHEMISTRY AND MOLECULAR BIOLOGY Revealing how life works Department of Biochemistry and Biophysics Oregon State University 2011 Agriculture and Life Sciences Building Corvallis, OR 97331 OSUBB 541-737-4511 biochem.oregonstate.edu College of Science Oregon State University 128 Kidder Hall Corvallis, OR 97331 541-737-4811 science.oregonstate.edu This publication will be made available in an accessible alternative format upon request. Please contact the College COLLEGE OF SCIENCE of Science at 541-737-4811 or [email protected]. ACADEMIC BROCHURE / 2020 Highlights • Solve problems at the What will you discover? intersection of biological and physical sciences with our The Department of Biochemistry and Biophysics offers nationally accredited program world-class faculty, a tradition of interdisciplinary in biochemistry, molecular and research, teaching excellence and extraordinary cellular biology, chemistry, laboratories to facilitate undergraduate learning. molecular genetics, physics and statistics. The department ranks high nationally and internationally in many research areas, including signal transduction, gene • Tailor your education to expression, epigenetics, metabolic regulation, structural specific career goals with three biology, and genetic code expansion technology. different options. The department offers two Bachelor of Science degrees, • Thrive in a smaller, supportive both accredited by the American Society for Biochemistry department within a world-class and Molecular Biology (ASBMB): research university. • Biochemistry and Molecular Biology (BMB) with • Pursue interdisciplinary research options in Advanced Molecular Biology, Computational projects with faculty across OSU. Molecular Biology, and Pre-medicine; • Participate in the Biochemistry • Biochemistry and Biophysics (BB) with options in Club for community, leadership Advanced Biophysics, Neuroscience, and Pre-medicine. -

1 a Primer on Molecular Biology
1APrimer on Molecular Biology Alexander Zien Modern molecular biology provides a rich source of challenging machine learning problems. This tutorial chapter aims to provide the necessary biological background knowledge required to communicate with biologists and to understand and properly formalize a number of most interesting problems in this application domain. The largest part of the chapter (its first section) is devoted to the cell as the basic unit of life. Four aspects of cells are reviewed in sequence: (1) the molecules that cells make use of (above all, proteins, RNA, and DNA); (2) the spatial organization of cells (“compartmentalization”); (3) the way cells produce proteins (“protein expression”); and (4) cellular communication and evolution (of cells and organisms). In the second section, an overview is provided of the most frequent measurement technologies, data types, and data sources. Finally, important open problems in the analysis of these data (bioinformatics challenges) are briefly outlined. 1.1 The Cell The basic unit of all (biological) life is the cell. A cell is basically a watery solution of certain molecules, surrounded by a lipid (fat) membrane. Typical sizes of cells range from 1 µm (bacteria) to 100 µm (plant cells). The most important properties Life of a living cell (and, in fact, of life itself) are the following: It consists of a set of molecules that is separated from the exterior (as a human being is separated from his or her surroundings). It has a metabolism, that is, it can take up nutrients and convert them into other molecules and usable energy. The cell uses nutrients to renew its constituents, to grow, and to drive its actions (just like a human does). -

Biology 302 Course Syllabus Cell and Molecular Biology Fall 2014
Biology 302 Course Syllabus Cell and Molecular Biology Fall 2014 Instructor: Dr. Karen K. Resendes Office Phone: 724-946-7211 Office: 222 Hoyt Science Center e-mail: [email protected] Course Description: The primary emphasis of this course will be the basic structure and function of the cell. We begin the semester by looking at the “smaller picture” – the basic types of organic molecules that contribute to cells. We then explore cellular processes by starting on the cell surface and working our way into the interior of the cell. General topics include membrane transport and signaling, gene expression, intracellular transport, structure and motility, energy conversions, and cell division. Laboratory exercises will reinforce many of concepts covered in lecture. This course assumes the student has learned and retained a basic understanding of the fundamental cellular concepts presented in Foundations I and II. Course Structure: Lecture: T, R 9:20AM – 11:00AM Laboratory: R 2:00PM – 5:00PM HSC 206 Hoyt 343 Office Hours: M and W 1PM-2:30 PM OR other times by appointment Simply email, call or ask me for a time that will work for you. The sole purpose of making an appointment is that I want to be sure that I will be in my office at the time you want to meet with me. Required: 1. Text: Alberts A., et al. Molecular Biology of the Cell. Fifth Edition. Garland Science Publishers. New York, NY. 2008 2.Composition-style, quad-lined laboratory notebook 3. Frequent visits to the Bio 302 coursepage on my.westminster for: -Laboratory Handouts - Primary research articles - Online quizzes -Detailed reading lists - Electronic Resources Course Objectives: This course is intended for those students majoring in biology, molecular biology, biochemistry, or neuroscience. -

Biochemistry and Molecular Biology (BMB) 1
Biochemistry and Molecular Biology (BMB) 1 BMB 372. Advanced Molecular Biology. (3 h) BIOCHEMISTRY AND Presents molecular mechanisms by which stored genetic information is expressed including the mechanisms for and regulation of gene MOLECULAR BIOLOGY (BMB) expression, protein synthesis, and genome editing. Emphasizes analysis and interpretation of experimental data from the primary literature. Also Interdisciplinary Major listed as BIO 372. P-BIO 213 and 214 and BIO 370/BMB 370/CHM 370 or BIO 265 and BIO 370/BMB 370/CHM 370. This interdisciplinary Bachelor of Science major, jointly offered by the Departments of Biology and Chemistry, provides a strong foundation BMB 372L. Advanced Molecular Biology Laboratory. (1.5 h) in biological chemistry and molecular biology, and related topics at Introduces modern methods of molecular biology to analyze and the interface of these two disciplines. The major is designed to build manipulate expression of genes and function of gene products. Also conceptual understanding and practical and critical thinking skills to listed as BIO 372L. P or C-BIO 372/CHM 372 or BMB 373/CHM 373. address current biological, biochemical, and biomedical challenges. A BMB 373. Biochemistry II. (3 h) required research experience spanning multiple semesters, culminating Examines the structure, function, and synthesis of proteins and nucleic in a senior project, will give students strong experimental skills and acids and includes advanced topics in biochemistry including catalytic provide insight into biochemical and molecular biological experimental mechanisms of enzymes and ribozymes, use of sequence and structure approaches and results that demonstrate the function of biological databases, and molecular basis of disease and drug action. -

Complete Nucleotide Sequence, Molecular Analysis and Genome Structure of Bacteriophage A118 of Listeria Monocytogenes : Implications for Phage Evolution
Molecular Microbiology (2000) 35(2), 324±340 Complete nucleotide sequence, molecular analysis and genome structure of bacteriophage A118 of Listeria monocytogenes : implications for phage evolution Martin J. Loessner,1* Ross B. Inman,2 Peter Lauer3 local, but sometimes extensive, similarities to a num- and Richard Calendar3 ber of phages spanning a broader phylogenetic range 1Institut fuÈr Mikrobiologie, FML Weihenstephan, of various low GC host bacteria, which implies rela- Technische UniversitaÈtMuÈnchen, Weihenstephaner tively recent exchange of genes or genetic modules. Berg 3, 85350 Freising, Germany. We have also identi®ed the A118 attachment site 2Institute for Molecular Virology, University of attP and the corresponding attB in Listeria monocyto- Wisconsin at Madison, 1525 Linden Drive, Madison, genes, and show that site-speci®c integration of the Wisconsin 53706, USA. A118 prophage by the A118 integrase occurs into a 3Department of Molecular and Cell Biology, host gene homologous to comK of Bacillus subtilis, University of California at Berkeley, 401 Barker Hall, an autoregulatory gene specifying the major compe- Berkeley, California 94720-3202, USA. tence transcription factor. Summary Introduction A118 is a temperate phage isolated from Listeria Listeria monocytogenes is a non-spore-forming, opportu- monocytogenes. In this study, we report the entire nistic Gram-positive pathogen, responsible for severe nucleotide sequence and structural analysis of its infections in both animals and humans, which is almost 40 834 bp DNA. Electron microscopic and enzymatic exclusively transmitted via contaminated food. Recurrent analyses revealed that the A118 genome is a linear, outbreaks of Listeriosis (CDC, 1998; Slutsker and Schuchat, circularly permuted, terminally redundant collection 1999) have emphasized the need for a better understand- of double-stranded DNA molecules. -

An Introduction to the Cell and Molecular Biology (CMB) Subprogram
An Introduction to the Cell and Molecular Biology (CMB) Subprogram SoLS Research Faculty Affiliated with the Cell and Molecular Biology (CMB) Graduate Subprogram • Andrew Andres • Nora Caberoy • Allen Gibbs • Mira Han • Allyson Hindle • Laurel Raftery • Martin Schiller • Jeffery Shen • Boo Shan Tseng • Kelly Tseng • Frank van Breukelen • Mo Weng •Helen Wing Required Courses for All Degrees • Biol 701—Ethics in Scientific Research (1 credit). Required class for all students in both degree programs. • Biol 790—Research Colloquium in Life Sciences Students may take this course for credit (1-2 credits/semester for a maximum of 9 credits toward the degree), but all students (including non-enrolled) must participate each semester. Core Courses for MS and PhD MS students must take at least ONE and PhD students must take at least THREE of the following Core Classes: • Biol 607—Molecular Biology (3 credits) • Biol 625—Genomics (3 credits) • Biol 645—Cell Physiology (3 credits) • Chem 772—Nucleic Acid Chemistry (3 credits) Elective Courses for MS and PhD • MS students must take TWO of the following electives. • PhD Students must take THREE of the following electives. Note: • For students whose research topic focuses predominantly on a cell biological topic, the advisory committee may choose a relevant elective course to replace one of the core courses. • Core courses (above) that are not being used to satisfy Core Requirements may be taken as Electives. • Elective courses must be approved in advance by the student’s Research Advisory Committee. • The Research Advisory Committee may require the student to take specific courses, depending on the person’s academic background and research objectives. -
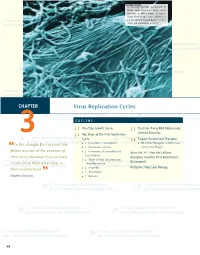
Virus Replication Cycles
© Jones and Bartlett Publishers. NOT FOR SALE OR DISTRIBUTION A scanning electron micrograph of Ebola virus particles. Ebola virus contains an RNA genome. It causes Ebola hemorrhagic fever, which is a severe and often fatal disease in hu- mans and nonhuman primates. CHAPTER Virus Replication Cycles OUTLINE 3.1 One-Step Growth Curves 3.3 The Error-Prone RNA Polymerases: 3 3.2 Key Steps of the Viral Replication Genetic Diversity Cycle 3.4 Targets for Antiviral Therapies In the struggle for survival, the ■ 1. Attachment (Adsorption) ■ RNA Virus Mutagens: A New Class “ ■ 2. Penetration (Entry) of Antiviral Drugs? fi ttest win out at the expense of ■ 3. Uncoating (Disassembly and Virus File 3-1: How Are Cellular Localization) their rivals because they succeed Receptors Used for Viral Attachment ■ 4. Types of Viral Genomes and Discovered? in adapting themselves best to Their Replication their environment. ■ 5. Assembly Refresher: Molecular Biology ” ■ 6. Maturation Charles Darwin ■ 7. Release 46 229329_CH03_046_069.indd9329_CH03_046_069.indd 4466 11/18/08/18/08 33:19:08:19:08 PPMM © Jones and Bartlett Publishers. NOT FOR SALE OR DISTRIBUTION CASE STUDY The campus day care was recently closed during the peak of the winter fl u season because many of the young children were sick with a lower respiratory tract infection. An email an- nouncement was sent to all students, faculty, and staff at the college that stated the closure was due to a metapneumovirus outbreak. The announcement briefed the campus com- munity with information about human metapneumonoviruses (hMPVs). The announcement stated that hMPV was a newly identifi ed respiratory tract pathogen discovered in the Netherlands in 2001. -

Space Biology Science Plan 2016 - 2025 Chapter X of the SLPSRA Integrated Research Plan
1 Space Biology Science Plan 2016 - 2025 Chapter X of the SLPSRA Integrated Research Plan Cell and Molecular Microbial Developmental Invertebrate Plant Vertebrate Space Biology Science Plan 2016-2025 May 11, 2016 – Page 2 Dedication A core team of authors began working on this plan for Space Biology research in mid-June 2015 at a retreat at Ames Research Center. The team that assembled at Ames that week included David Tomko, Ken Souza, Jeff Smith, Richard Mains, Kevin Sato, Howard Levine, Charlie Quincy, Aaron Mills and Amir Zeituni. Although others have contributed to our efforts over the past year, this team of authors bears principal re- sponsibility for its shape and content. During the past month Ken worked with me almost non-stop on the final polish, to insure as much as humanly possible that errors were eliminated and nothing was left out. His contribution to Space Biology is immeasurable, the Plan would not exist in its present form without his work, and we will sorely miss work- ing with him in the future. It is therefore with the greatest respect and admiration that we dedicate this Space Biology Plan to the memory of our dear friend, colleague and mentor Ken Souza, who worked tirelessly, well and hard with us on its genesis. – The Authors, March 23, 2016 3 Chapter X In SLPSRA Integrated Research Plan Space Biology Plan 2016-2025 I Background A. Introduction B. Congressional Direction to NASA C. Major NRC Decadal Survey Recommendations II. Summary of Progress since the 2010 Space Biology Science Plan. A. Goals of the 2010 Space Biology Science Plan and summary of progress III.