Nanoscale ARTICLE
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Defining Functional Interactions During Biogenesis of Epithelial Junctions
ARTICLE Received 11 Dec 2015 | Accepted 13 Oct 2016 | Published 6 Dec 2016 | Updated 5 Jan 2017 DOI: 10.1038/ncomms13542 OPEN Defining functional interactions during biogenesis of epithelial junctions J.C. Erasmus1,*, S. Bruche1,*,w, L. Pizarro1,2,*, N. Maimari1,3,*, T. Poggioli1,w, C. Tomlinson4,J.Lees5, I. Zalivina1,w, A. Wheeler1,w, A. Alberts6, A. Russo2 & V.M.M. Braga1 In spite of extensive recent progress, a comprehensive understanding of how actin cytoskeleton remodelling supports stable junctions remains to be established. Here we design a platform that integrates actin functions with optimized phenotypic clustering and identify new cytoskeletal proteins, their functional hierarchy and pathways that modulate E-cadherin adhesion. Depletion of EEF1A, an actin bundling protein, increases E-cadherin levels at junctions without a corresponding reinforcement of cell–cell contacts. This unexpected result reflects a more dynamic and mobile junctional actin in EEF1A-depleted cells. A partner for EEF1A in cadherin contact maintenance is the formin DIAPH2, which interacts with EEF1A. In contrast, depletion of either the endocytic regulator TRIP10 or the Rho GTPase activator VAV2 reduces E-cadherin levels at junctions. TRIP10 binds to and requires VAV2 function for its junctional localization. Overall, we present new conceptual insights on junction stabilization, which integrate known and novel pathways with impact for epithelial morphogenesis, homeostasis and diseases. 1 National Heart and Lung Institute, Faculty of Medicine, Imperial College London, London SW7 2AZ, UK. 2 Computing Department, Imperial College London, London SW7 2AZ, UK. 3 Bioengineering Department, Faculty of Engineering, Imperial College London, London SW7 2AZ, UK. 4 Department of Surgery & Cancer, Faculty of Medicine, Imperial College London, London SW7 2AZ, UK. -

Vocabulario De Morfoloxía, Anatomía E Citoloxía Veterinaria
Vocabulario de Morfoloxía, anatomía e citoloxía veterinaria (galego-español-inglés) Servizo de Normalización Lingüística Universidade de Santiago de Compostela COLECCIÓN VOCABULARIOS TEMÁTICOS N.º 4 SERVIZO DE NORMALIZACIÓN LINGÜÍSTICA Vocabulario de Morfoloxía, anatomía e citoloxía veterinaria (galego-español-inglés) 2008 UNIVERSIDADE DE SANTIAGO DE COMPOSTELA VOCABULARIO de morfoloxía, anatomía e citoloxía veterinaria : (galego-español- inglés) / coordinador Xusto A. Rodríguez Río, Servizo de Normalización Lingüística ; autores Matilde Lombardero Fernández ... [et al.]. – Santiago de Compostela : Universidade de Santiago de Compostela, Servizo de Publicacións e Intercambio Científico, 2008. – 369 p. ; 21 cm. – (Vocabularios temáticos ; 4). - D.L. C 2458-2008. – ISBN 978-84-9887-018-3 1.Medicina �������������������������������������������������������������������������veterinaria-Diccionarios�������������������������������������������������. 2.Galego (Lingua)-Glosarios, vocabularios, etc. políglotas. I.Lombardero Fernández, Matilde. II.Rodríguez Rio, Xusto A. coord. III. Universidade de Santiago de Compostela. Servizo de Normalización Lingüística, coord. IV.Universidade de Santiago de Compostela. Servizo de Publicacións e Intercambio Científico, ed. V.Serie. 591.4(038)=699=60=20 Coordinador Xusto A. Rodríguez Río (Área de Terminoloxía. Servizo de Normalización Lingüística. Universidade de Santiago de Compostela) Autoras/res Matilde Lombardero Fernández (doutora en Veterinaria e profesora do Departamento de Anatomía e Produción Animal. -

Plakoglobin Is Required for Effective Intermediate Filament Anchorage to Desmosomes Devrim Acehan1, Christopher Petzold1, Iwona Gumper2, David D
ORIGINAL ARTICLE Plakoglobin Is Required for Effective Intermediate Filament Anchorage to Desmosomes Devrim Acehan1, Christopher Petzold1, Iwona Gumper2, David D. Sabatini2, Eliane J. Mu¨ller3, Pamela Cowin2,4 and David L. Stokes1,2,5 Desmosomes are adhesive junctions that provide mechanical coupling between cells. Plakoglobin (PG) is a major component of the intracellular plaque that serves to connect transmembrane elements to the cytoskeleton. We have used electron tomography and immunolabeling to investigate the consequences of PG knockout on the molecular architecture of the intracellular plaque in cultured keratinocytes. Although knockout keratinocytes form substantial numbers of desmosome-like junctions and have a relatively normal intercellular distribution of desmosomal cadherins, their cytoplasmic plaques are sparse and anchoring of intermediate filaments is defective. In the knockout, b-catenin appears to substitute for PG in the clustering of cadherins, but is unable to recruit normal levels of plakophilin-1 and desmoplakin to the plaque. By comparing tomograms of wild type and knockout desmosomes, we have assigned particular densities to desmoplakin and described their interaction with intermediate filaments. Desmoplakin molecules are more extended in wild type than knockout desmosomes, as if intermediate filament connections produced tension within the plaque. On the basis of our observations, we propose a particular assembly sequence, beginning with cadherin clustering within the plasma membrane, followed by recruitment of plakophilin and desmoplakin to the plaque, and ending with anchoring of intermediate filaments, which represents the key to adhesive strength. Journal of Investigative Dermatology (2008) 128, 2665–2675; doi:10.1038/jid.2008.141; published online 22 May 2008 INTRODUCTION dense plaque that is further from the membrane and that Desmosomes are large macromolecular complexes that mediates the binding of intermediate filaments. -

Supplementary Table 1: Adhesion Genes Data Set
Supplementary Table 1: Adhesion genes data set PROBE Entrez Gene ID Celera Gene ID Gene_Symbol Gene_Name 160832 1 hCG201364.3 A1BG alpha-1-B glycoprotein 223658 1 hCG201364.3 A1BG alpha-1-B glycoprotein 212988 102 hCG40040.3 ADAM10 ADAM metallopeptidase domain 10 133411 4185 hCG28232.2 ADAM11 ADAM metallopeptidase domain 11 110695 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 195222 8038 hCG40937.4 ADAM12 ADAM metallopeptidase domain 12 (meltrin alpha) 165344 8751 hCG20021.3 ADAM15 ADAM metallopeptidase domain 15 (metargidin) 189065 6868 null ADAM17 ADAM metallopeptidase domain 17 (tumor necrosis factor, alpha, converting enzyme) 108119 8728 hCG15398.4 ADAM19 ADAM metallopeptidase domain 19 (meltrin beta) 117763 8748 hCG20675.3 ADAM20 ADAM metallopeptidase domain 20 126448 8747 hCG1785634.2 ADAM21 ADAM metallopeptidase domain 21 208981 8747 hCG1785634.2|hCG2042897 ADAM21 ADAM metallopeptidase domain 21 180903 53616 hCG17212.4 ADAM22 ADAM metallopeptidase domain 22 177272 8745 hCG1811623.1 ADAM23 ADAM metallopeptidase domain 23 102384 10863 hCG1818505.1 ADAM28 ADAM metallopeptidase domain 28 119968 11086 hCG1786734.2 ADAM29 ADAM metallopeptidase domain 29 205542 11085 hCG1997196.1 ADAM30 ADAM metallopeptidase domain 30 148417 80332 hCG39255.4 ADAM33 ADAM metallopeptidase domain 33 140492 8756 hCG1789002.2 ADAM7 ADAM metallopeptidase domain 7 122603 101 hCG1816947.1 ADAM8 ADAM metallopeptidase domain 8 183965 8754 hCG1996391 ADAM9 ADAM metallopeptidase domain 9 (meltrin gamma) 129974 27299 hCG15447.3 ADAMDEC1 ADAM-like, -

Cell Biology of Tight Junction Barrier Regulation and Mucosal Disease
Downloaded from http://cshperspectives.cshlp.org/ on October 1, 2021 - Published by Cold Spring Harbor Laboratory Press Cell Biology of Tight Junction Barrier Regulation and Mucosal Disease Aaron Buckley and Jerrold R. Turner Departments of Pathology and Medicine (Gastroenterology), Brigham and Women’s Hospital and Harvard Medical School, Boston, Massachusetts 02115 Correspondence: [email protected] Mucosal surfaces are lined by epithelial cells. In the intestine, the epithelium establishes a selectively permeable barrier that supports nutrient absorption and waste secretion while preventing intrusion by luminal materials. Intestinal epithelia therefore play a central role in regulating interactions between the mucosal immune system and luminal contents, which include dietary antigens, a diverse intestinal microbiome, and pathogens. The paracellular space is sealed by the tight junction, which is maintained by a complex network of protein interactions. Tight junction dysfunction has been linked to a variety of local and systemic diseases. Two molecularly and biophysically distinct pathways across the intestinal tight junc- tion are selectively and differentially regulated by inflammatory stimuli. This review discusses the mechanisms underlying these events, their impact on disease, and the potential of using these as paradigms for development of tight junction-targeted therapeutic interventions. ucosal surfaces and the epithelial cells that adherens). The tight junction is a selectively Mline them are present at sites where tissues permeable barrier that generally represents the interface directly with the external environment rate-limiting step of paracellular transport. The or internal compartments that are contiguous adherens junction and desmosome provide es- with the external environment. Examples in- sential adhesive and mechanical properties that clude the gastrointestinal tract, the pulmonary contribute to barrier function but do not seal tree, and the genitourinary tract. -

Hemidesmosomes Show Abnormal Association with the Keratin Filament Network in Junctional Forms of Epidermolysis Bullosa
View metadata, citation and similar papers at core.ac.uk brought to you by CORE provided by Elsevier - Publisher Connector Hemidesmosomes Show Abnormal Association with the Keratin Filament Network in Junctional Forms of Epidermolysis Bullosa James R. McMillan, John A. McGrath, Michael J. Tidman,* and Robin A. J. Eady Department of Cell Pathology, St John’s Institute of Dermatology, UMDS, St Thomas’s Hospital, London, U.K.; *Department of Dermatology, Royal Infirmary of Edinburgh, Edinburgh, U.K. Junctional epidermolysis bullosa is a group of hereditary respectively. In junctional epidermolysis bullosa with bullous disorders resulting from defects in several hemi- pyloric atresia (α6β4 abnormalities, n J 3) the values desmosome-anchoring filament components. Because were also reduced [41.8% K 7.0 (p < 0.001) and 44.5% hemidesmosomes are involved not only in keratinocyte- K 5.7 (p < 0.001), respectively]. In the non-Herlitz group extracellular matrix adherence, but also in normal (laminin 5 mutations, n J 3) the counts were 66.7% K anchorage of keratin intermediate filaments to the basal 7.1 (p > 0.05) and 70.5% K 8.5 (p < 0.05), and in keratinocyte membrane, we questioned whether this skin from patients with bullous pemphigoid antigen 2 intracellular function of hemidesmosomes was also per- mutations (n J 3) the counts were 54.3% K 13.8 (p < 0.01) turbed in junctional epidermolysis bullosa. We used and 57.1% K 13.9 (p < 0.01). In epidermolysis bullosa quantitative electron microscopic methods to assess cer- simplex associated with plectin mutations the values were tain morphologic features of hemidesmosome–keratin 31.9% K 8.9 (p < 0.001) for keratin intermediate filaments intermediate filaments interactions in skin from normal association and 39.9% K 7.1 (p < 0.001) for inner plaques. -

Talin-1 Is the Principal Platelet Rap1 Effector of Integrin Activation
Regular Article PLATELETS AND THROMBOPOIESIS Talin-1 is the principal platelet Rap1 effector of integrin activation 1,2, 3,4, 1, 1 1 1 1 Frederic Lagarrigue, * David S. Paul, * Alexandre R. Gingras, * Andrew J. Valadez, Hao Sun, Jenny Lin, Monica N. Cuevas, Downloaded from http://ashpublications.org/blood/article-pdf/136/10/1180/1756644/bloodbld2020005348.pdf by UNIV OF NC/ACQ SRVCS user on 20 October 2020 Jailal N. Ablack,1 Miguel Alejandro Lopez-Ramirez,1,5 Wolfgang Bergmeier,3,4 and Mark H. Ginsberg1 1Department of Medicine, University of California, San Diego, La Jolla, CA; 2Institut de Pharmacologie et de Biologie Structurale (IPBS), Centre National de la Recherche Scientifique (CNRS), Universite´ Paul Sabatier (UPS), Universite´ de Toulouse, Toulouse, France; 3UNC Blood Research Center and 4Department of Biochemistry and Biophysics, University of North Carolina at Chapel Hill, Chapel Hill, NC; and 5Department of Pharmacology, University of California, San Diego, La Jolla, CA KEY POINTS Ras-related protein 1 (Rap1) is a major convergence point of the platelet-signaling path- ways that result in talin-1 binding to the integrin b cytoplasmic domain and consequent l Blockade of Rap1 binding to talin-1 F0 integrin activation, platelet aggregation, and effective hemostasis. The nature of the and F1 domains connection between Rap1 and talin-1 in integrin activation is an important remaining gap in phenocopies the our understanding of this process. Previous work identified a low-affinity Rap1-binding site defects in integrin in the talin-1 F0 domain that makes a small contribution to integrin activation in platelets. activation observed in platelets lacking We recently identified an additional Rap1-binding site in the talin-1 F1 domain that makes a Rap1a/b. -

Nomina Histologica Veterinaria, First Edition
NOMINA HISTOLOGICA VETERINARIA Submitted by the International Committee on Veterinary Histological Nomenclature (ICVHN) to the World Association of Veterinary Anatomists Published on the website of the World Association of Veterinary Anatomists www.wava-amav.org 2017 CONTENTS Introduction i Principles of term construction in N.H.V. iii Cytologia – Cytology 1 Textus epithelialis – Epithelial tissue 10 Textus connectivus – Connective tissue 13 Sanguis et Lympha – Blood and Lymph 17 Textus muscularis – Muscle tissue 19 Textus nervosus – Nerve tissue 20 Splanchnologia – Viscera 23 Systema digestorium – Digestive system 24 Systema respiratorium – Respiratory system 32 Systema urinarium – Urinary system 35 Organa genitalia masculina – Male genital system 38 Organa genitalia feminina – Female genital system 42 Systema endocrinum – Endocrine system 45 Systema cardiovasculare et lymphaticum [Angiologia] – Cardiovascular and lymphatic system 47 Systema nervosum – Nervous system 52 Receptores sensorii et Organa sensuum – Sensory receptors and Sense organs 58 Integumentum – Integument 64 INTRODUCTION The preparations leading to the publication of the present first edition of the Nomina Histologica Veterinaria has a long history spanning more than 50 years. Under the auspices of the World Association of Veterinary Anatomists (W.A.V.A.), the International Committee on Veterinary Anatomical Nomenclature (I.C.V.A.N.) appointed in Giessen, 1965, a Subcommittee on Histology and Embryology which started a working relation with the Subcommittee on Histology of the former International Anatomical Nomenclature Committee. In Mexico City, 1971, this Subcommittee presented a document entitled Nomina Histologica Veterinaria: A Working Draft as a basis for the continued work of the newly-appointed Subcommittee on Histological Nomenclature. This resulted in the editing of the Nomina Histologica Veterinaria: A Working Draft II (Toulouse, 1974), followed by preparations for publication of a Nomina Histologica Veterinaria. -

Molecular Organization of the Desmosome As Revealed by Direct Stochastic Optical Reconstruction Microscopy Sara N
© 2016. Published by The Company of Biologists Ltd | Journal of Cell Science (2016) 129, 2897-2904 doi:10.1242/jcs.185785 SHORT REPORT Molecular organization of the desmosome as revealed by direct stochastic optical reconstruction microscopy Sara N. Stahley1, Emily I. Bartle1, Claire E. Atkinson2, Andrew P. Kowalczyk1,3 and Alexa L. Mattheyses1,* ABSTRACT plakoglobin and plakophilin, and the plakin family member Desmosomes are macromolecular junctions responsible for providing desmoplakin contribute to the intracellular plaque (Fig. 1A). strong cell–cell adhesion. Because of their size and molecular Plaque ultrastructure is characterized by two electron-dense complexity, the precise ultrastructural organization of desmosomes regions: the plasma-membrane-proximal outer dense plaque and is challenging to study. Here, we used direct stochastic optical the inner dense plaque (Desai et al., 2009; Farquhar and Palade, reconstruction microscopy (dSTORM) to resolve individual plaque 1963; Stokes, 2007). The cadherin cytoplasmic tails bind to proteins pairs for inner and outer dense plaque proteins. Analysis methods in the outer dense plaque whereas the C-terminus of desmoplakin based on desmosomal mirror symmetry were developed to measure binds to intermediate filaments in the inner dense plaque. This plaque-to-plaque distances and create an integrated map. We tethers the desmosome to the intermediate filament cytoskeleton, quantified the organization of desmoglein 3, plakoglobin and establishing an integrated adhesive network (Bornslaeger et al., desmoplakin (N-terminal, rod and C-terminal domains) in primary 1996; Harmon and Green, 2013). human keratinocytes. Longer desmosome lengths correlated with Many desmosomal protein interactions have been characterized increasing plaque-to-plaque distance, suggesting that desmoplakin is by biochemical studies (Bass-Zubek and Green, 2007; Green and arranged with its long axis at an angle within the plaque. -

Adherens Junctions, Desmosomes and Tight Junctions in Epidermal Barrier Function Johanna M
14 The Open Dermatology Journal, 2010, 4, 14-20 Open Access Adherens Junctions, Desmosomes and Tight Junctions in Epidermal Barrier Function Johanna M. Brandner1,§, Marek Haftek*,2,§ and Carien M. Niessen3,§ 1Department of Dermatology and Venerology, University Hospital Hamburg-Eppendorf, Hamburg, Germany 2University of Lyon, EA4169 Normal and Pathological Functions of Skin Barrier, E. Herriot Hospital, Lyon, France 3Department of Dermatology, Center for Molecular Medicine, Cologne Excellence Cluster on Cellular Stress Responses in Aging-Associated Diseases (CECAD), University of Cologne, Germany Abstract: The skin is an indispensable barrier which protects the body from the uncontrolled loss of water and solutes as well as from chemical and physical assaults and the invasion of pathogens. In recent years several studies have suggested an important role of intercellular junctions for the barrier function of the epidermis. In this review we summarize our knowledge of the impact of adherens junctions, (corneo)-desmosomes and tight junctions on barrier function of the skin. Keywords: Cadherins, catenins, claudins, cell polarity, stratum corneum, skin diseases. INTRODUCTION ADHERENS JUNCTIONS The stratifying epidermis of the skin physically separates Adherens junctions are intercellular structures that couple the organism from its environment and serves as its first line intercellular adhesion to the cytoskeleton thereby creating a of structural and functional defense against dehydration, transcellular network that coordinate the behavior of a chemical substances, physical insults and micro-organisms. population of cells. Adherens junctions are dynamic entities The living cell layers of the epidermis are crucial in the and also function as signal platforms that regulate formation and maintenance of the barrier on two different cytoskeletal dynamics and cell polarity. -

Loss of Mouse Cardiomyocyte Talin-1 and Talin-2 Leads to Β-1 Integrin
Loss of mouse cardiomyocyte talin-1 and talin-2 leads PNAS PLUS to β-1 integrin reduction, costameric instability, and dilated cardiomyopathy Ana Maria Mansoa,b,1, Hideshi Okadaa,b, Francesca M. Sakamotoa, Emily Morenoa, Susan J. Monkleyc, Ruixia Lia, David R. Critchleyc, and Robert S. Rossa,b,1 aDivision of Cardiology, Department of Medicine, University of California at San Diego School of Medicine, La Jolla, CA 92093; bCardiology Section, Department of Medicine, Veterans Administration Healthcare, San Diego, CA 92161; and cDepartment of Molecular Cell Biology, University of Leicester, Leicester LE1 9HN, United Kingdom Edited by Kevin P. Campbell, Howard Hughes Medical Institute, University of Iowa, Iowa City, IA, and approved May 30, 2017 (received for review January 26, 2017) Continuous contraction–relaxation cycles of the heart require ognized as key mechanotransducers, converting mechanical per- strong and stable connections of cardiac myocytes (CMs) with turbations to biochemical signals (5, 6). the extracellular matrix (ECM) to preserve sarcolemmal integrity. The complex of proteins organized by integrins has been most CM attachment to the ECM is mediated by integrin complexes commonly termed focal adhesions (FA) by studies performed in localized at the muscle adhesion sites termed costameres. The cells such as fibroblasts in a 2D environment. It is recognized that ubiquitously expressed cytoskeletal protein talin (Tln) is a compo- this structure is important for organizing and regulating the me- nent of muscle costameres that links integrins ultimately to the chanical and signaling events that occur upon cellular adhesion to sarcomere. There are two talin genes, Tln1 and Tln2. Here, we ECM (7, 8). -
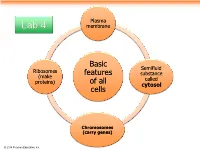
Basic Features of All Cells
Plasma Lab 4 membrane Basic Semifluid Ribosomes features substance (make called proteins) of all cytosol cells Chromosomes (carry genes) © 2014 Pearson Education, Inc. Prokaryotic cells are characterized by having . No nucleus . DNA in an unbound region called the nucleoid . No membrane-bound organelles . Cytoplasm bound by the plasma membrane © 2014 Pearson Education, Inc. Eukaryotic cells are characterized by having • DNA in a nucleus that is bounded by a membranous nuclear envelope • Membrane-bound organelles • Cytoplasm in the region between the plasma membrane and nucleus Eukaryotic cells are generally much larger than prokaryotic cells © 2014 Pearson Education, Inc. Figure 6.8a ENDOPLASMIC RETICULUM (ER) Nuclear envelope Rough ER Smooth ER Nucleolus NUCLEUS Flagellum Chromatin Centrosome Plasma membrane CYTOSKELETON: Microfilaments Intermediate filaments Microtubules Ribosomes Microvilli Golgi apparatus Peroxisome Lysosome Mitochondrion © 2014 Pearson Education, Inc. Figure 6.8b Nuclear envelope NUCLEUS Nucleolus Rough ER Chromatin Smooth ER Ribosomes Golgi Central vacuole apparatus Microfilaments CYTOSKELETON Microtubules Mitochondrion Peroxisome Plasma Chloroplast membrane Cell wall Plasmodesmata Wall of adjacent cell © 2014 Pearson Education, Inc. © 2014 Pearson Education, Inc. © 2014 Pearson Education, Inc. © 2014 Pearson Education, Inc. Table 6.1 © 2014 Pearson Education, Inc. Cell Walls of Plants The cell wall is an extracellular structure that distinguishes plant cells from animal cells Plant cell walls Prokaryotes, are made