NMN Reference Station Network Doing New Things in New Places – South Australian Gulfs
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

§4-71-6.5 LIST of CONDITIONALLY APPROVED ANIMALS November
§4-71-6.5 LIST OF CONDITIONALLY APPROVED ANIMALS November 28, 2006 SCIENTIFIC NAME COMMON NAME INVERTEBRATES PHYLUM Annelida CLASS Oligochaeta ORDER Plesiopora FAMILY Tubificidae Tubifex (all species in genus) worm, tubifex PHYLUM Arthropoda CLASS Crustacea ORDER Anostraca FAMILY Artemiidae Artemia (all species in genus) shrimp, brine ORDER Cladocera FAMILY Daphnidae Daphnia (all species in genus) flea, water ORDER Decapoda FAMILY Atelecyclidae Erimacrus isenbeckii crab, horsehair FAMILY Cancridae Cancer antennarius crab, California rock Cancer anthonyi crab, yellowstone Cancer borealis crab, Jonah Cancer magister crab, dungeness Cancer productus crab, rock (red) FAMILY Geryonidae Geryon affinis crab, golden FAMILY Lithodidae Paralithodes camtschatica crab, Alaskan king FAMILY Majidae Chionocetes bairdi crab, snow Chionocetes opilio crab, snow 1 CONDITIONAL ANIMAL LIST §4-71-6.5 SCIENTIFIC NAME COMMON NAME Chionocetes tanneri crab, snow FAMILY Nephropidae Homarus (all species in genus) lobster, true FAMILY Palaemonidae Macrobrachium lar shrimp, freshwater Macrobrachium rosenbergi prawn, giant long-legged FAMILY Palinuridae Jasus (all species in genus) crayfish, saltwater; lobster Panulirus argus lobster, Atlantic spiny Panulirus longipes femoristriga crayfish, saltwater Panulirus pencillatus lobster, spiny FAMILY Portunidae Callinectes sapidus crab, blue Scylla serrata crab, Samoan; serrate, swimming FAMILY Raninidae Ranina ranina crab, spanner; red frog, Hawaiian CLASS Insecta ORDER Coleoptera FAMILY Tenebrionidae Tenebrio molitor mealworm, -

An Investigation on Fishes of Bandirma Bay (Sea of Marmara)
BAÜ Fen Bil. Enst. Dergisi (2004).6.2 AN INVESTIGATION ON FISHES OF BANDIRMA BAY (SEA OF MARMARA) Hatice TORCU KOÇ University of Balikesir, Faculty of Science and Arts, Department of Hydrobiology, 10100, Balikesir, Turkey ABSTRACT This investigation was carried out for the determination of fish species living in Bandırma Bay (Sea of Marmara). Morphometric and meristic characters of of fishes caught by trawl and various nets in Bandırma Bay in the years of 1998-1999 were examined and some morphological, ecological properties, and local names of 34 determined species are given. Key Words: Fish Species, Systematic, Bandırma Bay BANDIRMA KÖRFEZİ (MARMARA DENİZİ) BALIKLARI ÜZERİNE BİR ARAŞTIRMA ÖZET Bu araştırma Bandırma Körfezi (Marmara Denizi)’nde yaşayan balık türlerini belirlemek amacıyla yapılmıştır. 1998-1999 yılları arasında körfez içinde trol ve çeşitli ağlar ile yakalanan balıkların morfometrik ve meristik karakterleri incelenmiş ve saptanan 34 türün bazı morfolojik, ekolojik özellikleri, ve yerel isimleri verilmiştir. Anahtar Kelimeler: Balık türleri, Sistematik, Bandırma Körfezi 1. INTRODUCTION Research on the sea fauna along the coasts of Turkey was initiated by foreign researchers at the begining of the 20th century and entered an intensive stage with Turkish researchers in the 1940s. However, the fish fauna of Turkish seas has still not been fully determined. Of these researchers, Tortonese (1) listed 300 species. Papaconstantinou and Tsimenids (2) listed 33 species. Papaconstantinou (3) listed the most of 447 species for Aegean Sea. Slastenenko (4) listed 200 species for Sea of Marmara and 189 species for Black Sea. Tortonese (1) reported 540 fish species in whole of Mediterranean. Demetropoulos and Neocleous (5) gave a list of fishes for Cyprus area. -

Fishes of Terengganu East Coast of Malay Peninsula, Malaysia Ii Iii
i Fishes of Terengganu East coast of Malay Peninsula, Malaysia ii iii Edited by Mizuki Matsunuma, Hiroyuki Motomura, Keiichi Matsuura, Noor Azhar M. Shazili and Mohd Azmi Ambak Photographed by Masatoshi Meguro and Mizuki Matsunuma iv Copy Right © 2011 by the National Museum of Nature and Science, Universiti Malaysia Terengganu and Kagoshima University Museum All rights reserved. No part of this publication may be reproduced or transmitted in any form or by any means without prior written permission from the publisher. Copyrights of the specimen photographs are held by the Kagoshima Uni- versity Museum. For bibliographic purposes this book should be cited as follows: Matsunuma, M., H. Motomura, K. Matsuura, N. A. M. Shazili and M. A. Ambak (eds.). 2011 (Nov.). Fishes of Terengganu – east coast of Malay Peninsula, Malaysia. National Museum of Nature and Science, Universiti Malaysia Terengganu and Kagoshima University Museum, ix + 251 pages. ISBN 978-4-87803-036-9 Corresponding editor: Hiroyuki Motomura (e-mail: [email protected]) v Preface Tropical seas in Southeast Asian countries are well known for their rich fish diversity found in various environments such as beautiful coral reefs, mud flats, sandy beaches, mangroves, and estuaries around river mouths. The South China Sea is a major water body containing a large and diverse fish fauna. However, many areas of the South China Sea, particularly in Malaysia and Vietnam, have been poorly studied in terms of fish taxonomy and diversity. Local fish scientists and students have frequently faced difficulty when try- ing to identify fishes in their home countries. During the International Training Program of the Japan Society for Promotion of Science (ITP of JSPS), two graduate students of Kagoshima University, Mr. -

Catch and Effort Data
Chapter 11: The Inland Waters River Fishery Information System CHAPTER 11: INLAND WATERS RIVER FISHERY INFORMATION SYSTEM 11.1 Background The Inland Waters River fishery is a multi-species, multi-gear fishery encompassing the South Australian sector of the River Murray and its backwaters (Map 11.1). Historically, the fishery was based on harvesting Murray cod (Maccullochella peelii peelii), golden perch (Macquaria ambigua), and bony bream (Nematalosa erebi). The fishery was restructured in July 2003 to a non-native dominate fishery and the commercial fishing for significant native species is now prohibited. The fishery is now based predominantly on the taking European carp (Cyprinus carpio) and redfin perch (Perca fluviatilis). There are a total of 6 licence holders who operate within the fishery. Prior to the restructure 30 license holders fished the river, each with a designated reach. Up to December 2007 SARDI production tables hold in excess of 270,000 daily records. Data is held from 1984/85 to current and is continually being updated. Refer to Figure 11.3 for an entity relationship diagram of the inland waters system that encompasses the River Fishery. South Australian Aquatic Sciences: Information Systems and Database Support Program SARDI Aquatic Sciences Information Systems Quality Assurance and Data Integrity Report (June 2009) 81 Chapter 11: The Inland Waters River Fishery Information System Map 11.1: The inland waters river fishery – River Murray area designations. South Australian Aquatic Sciences: Information Systems and Database Support Program SARDI Aquatic Sciences Information Systems Quality Assurance and Data Integrity Report (June 2009) 82 Chapter 11: The Inland Waters River Fishery Information System 11.2 Research Logbook Information Each licence holder is required to submit a daily fishing return by the 15th day of each month detailing their fishing activities for the previous month (Figure 11.1). -

New Zealand Fishes a Field Guide to Common Species Caught by Bottom, Midwater, and Surface Fishing Cover Photos: Top – Kingfish (Seriola Lalandi), Malcolm Francis
New Zealand fishes A field guide to common species caught by bottom, midwater, and surface fishing Cover photos: Top – Kingfish (Seriola lalandi), Malcolm Francis. Top left – Snapper (Chrysophrys auratus), Malcolm Francis. Centre – Catch of hoki (Macruronus novaezelandiae), Neil Bagley (NIWA). Bottom left – Jack mackerel (Trachurus sp.), Malcolm Francis. Bottom – Orange roughy (Hoplostethus atlanticus), NIWA. New Zealand fishes A field guide to common species caught by bottom, midwater, and surface fishing New Zealand Aquatic Environment and Biodiversity Report No: 208 Prepared for Fisheries New Zealand by P. J. McMillan M. P. Francis G. D. James L. J. Paul P. Marriott E. J. Mackay B. A. Wood D. W. Stevens L. H. Griggs S. J. Baird C. D. Roberts‡ A. L. Stewart‡ C. D. Struthers‡ J. E. Robbins NIWA, Private Bag 14901, Wellington 6241 ‡ Museum of New Zealand Te Papa Tongarewa, PO Box 467, Wellington, 6011Wellington ISSN 1176-9440 (print) ISSN 1179-6480 (online) ISBN 978-1-98-859425-5 (print) ISBN 978-1-98-859426-2 (online) 2019 Disclaimer While every effort was made to ensure the information in this publication is accurate, Fisheries New Zealand does not accept any responsibility or liability for error of fact, omission, interpretation or opinion that may be present, nor for the consequences of any decisions based on this information. Requests for further copies should be directed to: Publications Logistics Officer Ministry for Primary Industries PO Box 2526 WELLINGTON 6140 Email: [email protected] Telephone: 0800 00 83 33 Facsimile: 04-894 0300 This publication is also available on the Ministry for Primary Industries website at http://www.mpi.govt.nz/news-and-resources/publications/ A higher resolution (larger) PDF of this guide is also available by application to: [email protected] Citation: McMillan, P.J.; Francis, M.P.; James, G.D.; Paul, L.J.; Marriott, P.; Mackay, E.; Wood, B.A.; Stevens, D.W.; Griggs, L.H.; Baird, S.J.; Roberts, C.D.; Stewart, A.L.; Struthers, C.D.; Robbins, J.E. -
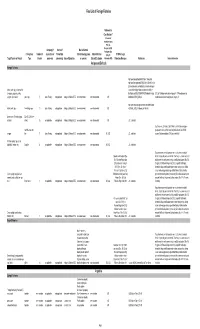
2018 Final LOFF W/ Ref and Detailed Info
Final List of Foreign Fisheries Rationale for Classification ** (Presence of mortality or injury (P/A), Co- Occurrence (C/O), Company (if Source of Marine Mammal Analogous Gear Fishery/Gear Number of aquaculture or Product (for Interactions (by group Marine Mammal (A/G), No RFMO or Legal Target Species or Product Type Vessels processor) processing) Area of Operation or species) Bycatch Estimates Information (N/I)) Protection Measures References Detailed Information Antigua and Barbuda Exempt Fisheries http://www.fao.org/fi/oldsite/FCP/en/ATG/body.htm http://www.fao.org/docrep/006/y5402e/y5402e06.htm,ht tp://www.tradeboss.com/default.cgi/action/viewcompan lobster, rock, spiny, demersal fish ies/searchterm/spiny+lobster/searchtermcondition/1/ , (snappers, groupers, grunts, ftp://ftp.fao.org/fi/DOCUMENT/IPOAS/national/Antigua U.S. LoF Caribbean spiny lobster trap/ pot >197 None documented, surgeonfish), flounder pots, traps 74 Lewis Fishing not applicable Antigua & Barbuda EEZ none documented none documented A/G AndBarbuda/NPOA_IUU.pdf Caribbean mixed species trap/pot are category III http://www.nmfs.noaa.gov/pr/interactions/fisheries/tabl lobster, rock, spiny free diving, loops 19 Lewis Fishing not applicable Antigua & Barbuda EEZ none documented none documented A/G e2/Atlantic_GOM_Caribbean_shellfish.html Queen conch (Strombus gigas), Dive (SCUBA & free molluscs diving) 25 not applicable not applicable Antigua & Barbuda EEZ none documented none documented A/G U.S. trade data Southeastern U.S. Atlantic, Gulf of Mexico, and Caribbean snapper- handline, hook and grouper and other reef fish bottom longline/hook-and-line/ >5,000 snapper line 71 Lewis Fishing not applicable Antigua & Barbuda EEZ none documented none documented N/I, A/G U.S. -

Targeted Review of Biological and Ecological Information from Fisheries Research in the South East Marine Region
TARGETED REVIEW OF BIOLOGICAL AND ECOLOGICAL INFORMATION FROM FISHERIES RESEARCH IN THE SOUTH EAST MARINE REGION FINAL REPORT B. D. Bruce, R. Bradford, R. Daley, M. Green and K. Phillips December 2002 Client: National Oceans Office Targeted review of biological and ecological information from fisheries research in the South East Marine Region Final Report B. D. Bruce, R. Bradford, R. Daley M. Green and K. Phillips* CSIRO Marine Research, Hobart * National Oceans Office December 2002 2 Table of Contents: Table of Contents:...................................................................................................................................3 Introduction.............................................................................................................................................5 Objective of review.............................................................................................................................5 Structure of review..............................................................................................................................5 Format.................................................................................................................................................6 General ecological/biological issues and uncertainties for the South East Marine Region ....................9 Specific fishery and key species accounts ............................................................................................10 South East Fishery (SEF) including the South East Trawl -

Ryan KL, Wise BS, Hall NG, Pollock KH, Sulin EH, Gaughan DJ (2013)
Fisheries Research Report No. 249, 2013 An integrated system to survey boat-based recreational fishing in Western Australia 2011/12 K.L. Ryan, B.S. Wise, N.G. Hall, K.H. Pollock, E.H. Sulin and D.J. Gaughan Fisheries Research Division Western Australian Fisheries and Marine Research Laboratories PO Box 20 NORTH BEACH, Western Australia 6920 Correct citation: Ryan KL, Wise BS, Hall NG, Pollock KH, Sulin EH, Gaughan DJ (2013). An integrated system to survey boat- based recreational fishing in Western Australia 2011/12. Fisheries Research Report No. 249, Department of Fisheries, Western Australia. 168pp. Enquiries: WA Fisheries and Marine Research Laboratories, PO Box 20, North Beach, WA 6920 Tel: +61 8 9203 0111 Email: [email protected] Website: www.fish.wa.gov.au ABN: 55 689 794 771 A complete list of Fisheries Research Reports is available online at www.fish.wa.gov.au © Department of Fisheries, Western Australia. September 2013. ISSN: 1035 - 4549 ISBN: 978-1-921845-71-3 ii Fisheries Research Report [Western Australia] No. 249, 2013 Contents 1.0 Introduction .................................................................................................................. 3 1.1 Importance of recreational fishing in WA .............................................................. 3 1.2 Need for recreational fishing information .............................................................. 3 1.3 Recreational fishing surveys in Australia ............................................................... 4 1.4 Recreational fishing surveys -

Redescription of Sphenanthias Whiteheadi Talwar (Perciformes: Cepolidae) with DNA Barcodes from the Southern Coasts of India
Zootaxa 3098: 64–68 (2011) ISSN 1175-5326 (print edition) www.mapress.com/zootaxa/ Article ZOOTAXA Copyright © 2011 · Magnolia Press ISSN 1175-5334 (online edition) Redescription of Sphenanthias whiteheadi Talwar (Perciformes: Cepolidae) with DNA barcodes from the southern coasts of India K.K. BINEESH1,3, K. A. SAJEELA2, K.V. AKHILESH1, N. G. K. PILLAI1 & E.M. ABDUSSAMAD1 1Central Marine Fisheries Research Institute (CMFRI), P. B. No. 1603, Ernakulam North P.O., Kochi - 683 018, Kerala, India 2National Bureau of Fish Genetic Resources (NBFGR), Cochin Unit, CMFRI Campus, P.B.No.1603, Ernakulam North, P.O., Kochi- 682 018, Kerala, India 3Corresponding author. E-mail: [email protected] Abstract A very rare bandfish, Sphenanthias whiteheadi Talwar 1973, is re-discovered and described from the southwest and south- east coasts of India for the first time after its original description and the rarity of the fish is challenged. A mitochondrial COI barcode sequence was generated for the specimen. Key words: Sphenanthias whiteheadi, Cepolidae, India Introduction The bandfishes of the family Cepolidae (Perciformes) are known from all tropical and subtropical waters and com- prise 22 valid species in 4 genera worldwide (Eschmeyer & Fong, 2011). Members of the genus Sphenanthias can be differentiated from the similar Owstonia in having lateral lines separate and not forming loops in front of dorsal fins (Smith Vaniz, 2001; Liao et al., 2009). The genus Sphenanthias is represented by only one valid species in the Arabian Sea (Manilo & Bogorodsky, 2003), Sphenanthias whiteheadi described by Talwar from four specimens collected from southwest coast of India off Quilon at 300 m (Talwar, 1973). -

Resource Assessments for Multi-Species Fisheries In
CM 2007/O:11 Resource assessments for multi-species fisheries in NSW, Australia: qualitative status determination using life history characteristics, empirical indicators and expert review. James Scandol and Kevin Rowling Wild Fisheries Program; Systems Research Branch Division of Science and Research; NSW Department of Primary Industries Cronulla Fisheries Centre; PO Box 21, Cronulla NSW 2230 Not to be cited without prior reference to the authors Abstract As the scope of fisheries management continues to broaden, there is increased pressure on scientific assessment processes to consider a greater number of species. This expanded list of species will inevitably include those with a range of life-history strategies, heterogeneous sources of information, and diverse stakeholder values. When faced with this challenge in New South Wales (Australia), the Department of Primary Industries developed systems and processes that scaled efficiently as the number of species requiring consideration increased. The key aspects of our approach are: • A qualitative determination of exploitation status based upon expert review. The current categories are: growth/recruitment overfished, fully fished, moderately fished, lightly fished, undefined and uncertain. • Application of management rules that require a Species Recovery Program should any species be determined to be overfished. • An emphasis on readily calculated empirical indicators (such as catch, catch-rates, length and age composition), rather than model-based estimates of biomass or fishing mortality. • Use of databases and electronic reporting systems to calculate empirical indicators based upon specified rules. Currently, there are 90 species considered with around six scientific and technical staff. These species were identified by commercial fishers as the important species from five multi-species and multi- method input-controlled fisheries. -

15.2 Sand Islands and Shoals
15 Islands 15.2 Sand Islands and Shoals Figure 15.1: (A) Aerial view of Troubridge Island and surrounding Troubridge Shoals: (c) Coastal Protection Branch, DEWNR. (B). Troubridge Island: (c) W. Bonham, Lighthouses of Australia. Asset Sand Islands and Shoals Description A crest of sand which rises above water level from a broad marine sand bank, forming an unstable sand island - Troubridge Island - which changes shape and size over time. The island is about 5m high at high tide, and about 2 hectares in area when inundated, but considerable larger at low tide. The island is surrounded by shallow sand embankments (Troubridge Shoals). Examples of Key Little Penguin, Black-faced Cormorant, Crested Tern and other breeding sea Species birds (numerous species) migratory wading birds (numerous species) abundant sand-dwelling invertebrates - food sources for fish and wading birds Pink Snapper King George whiting and school whiting syngnathid fishes (e.g. seahorses, pipefishes) sponges (forming “sponge gardens”, on consolidated sand) cowries; volutes and other specimen shells Knobby Argonaut (‘paper nautilus’ octopus) giant spider crab southern calamari Main Location Troubridge Island (and shallow sandbanks to the west - Troubridge Shoals) Notes Troubridge Island Conservation Park (approx. 260 hectares) was declared in 1982, and extended in 1986, partly to protect major breeding colonies of several seabird species, and provide protection for an important feeding ground used by migratory wading birds, listed under international treaties. Oceanography At the bottom of Gulf St Vincent, off the eastern “heel” of Yorke Peninsula, waters less than 20m occur up to 10km from shore. The oceanographic conditions have led to a long-term build-up of sand in some areas, including the creation of Troubridge Island, a sand island about 7km east of Sultana Point. -

Supplementary Material Figure S1 Mean Normalised Coastal Larval Fish
Supplementary Material Figure S1 Mean normalised coastal larval fish abundance among sampling projects from the Australian Integrated Marine Observing System (IMOS) Larval Fish Database (Smith et al., 2018). As projects were discrete periods of time, this also shows a relative stability in coastal larval abundance through time. Projects include two RV Franklin voyages (1994 & 1998), One voyage on the RV Kamala (1989-1990), two voyages on the RV Southern Surveyor (2004, 2010), one voyage on the RV Investigator (2015) and the ongoing IMOS Larval Fish monitoring program (shown as NIMO_NRS; 2014 – 2016). Figure S2 Temporal trends in total annual catch (‘000 kg) for four species from caught in gillnet fisheries on eight estuaries from July 1997 to June 2007. Note the varying scale on the y-axis. Figure S3 Temporal trends in total annual fishing effort (days) for gillnet fisheries on eight estuaries from July 1997 to June 2007. Note the log10 y-axis. Figure S4 Temporal trends in annual Catch-Per-Unit-Effort (CPUE; kg day-1) for four fish species caught in gillnet fisheries on eight estuaries from July 1997 to June 2007. Note the varying scale on the y-axis. Figure S5.1 Diagnostic plots of the parameters in the 14 day coastal larval fish Bayesian mixed model. Left hand panels show the distribution of the posterior estimates from 10,000 iterations. Right hand panels show the trace plots for the 4 chains post warm-up. Continued over the page. Figure S5.2 Diagnostic plots of the parameters in the 14 day winds coastal larval fish Bayesian mixed model.