2 Expression Is a Strong and Independent Predictor Of
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Nuclear Import Protein KPNA7 and Its Cargos Acta Universitatis Tamperensis 2346
ELISA VUORINEN Nuclear Import Protein KPNA7 and its Cargos ELISA Acta Universitatis Tamperensis 2346 ELISA VUORINEN Nuclear Import Protein KPNA7 and its Cargos Diverse roles in the regulation of cancer cell growth, mitosis and nuclear morphology AUT 2346 AUT ELISA VUORINEN Nuclear Import Protein KPNA7 and its Cargos Diverse roles in the regulation of cancer cell growth, mitosis and nuclear morphology ACADEMIC DISSERTATION To be presented, with the permission of the Faculty Council of the Faculty of Medicine and Life Sciences of the University of Tampere, for public discussion in the auditorium F114 of the Arvo building, Arvo Ylpön katu 34, Tampere, on 9 February 2018, at 12 o’clock. UNIVERSITY OF TAMPERE ELISA VUORINEN Nuclear Import Protein KPNA7 and its Cargos Diverse roles in the regulation of cancer cell growth, mitosis and nuclear morphology Acta Universitatis Tamperensis 2346 Tampere University Press Tampere 2018 ACADEMIC DISSERTATION University of Tampere, Faculty of Medicine and Life Sciences Finland Supervised by Reviewed by Professor Anne Kallioniemi Docent Pia Vahteristo University of Tampere University of Helsinki Finland Finland Docent Maria Vartiainen University of Helsinki Finland The originality of this thesis has been checked using the Turnitin OriginalityCheck service in accordance with the quality management system of the University of Tampere. Copyright ©2018 Tampere University Press and the author Cover design by Mikko Reinikka Acta Universitatis Tamperensis 2346 Acta Electronica Universitatis Tamperensis 1851 ISBN 978-952-03-0641-0 (print) ISBN 978-952-03-0642-7 (pdf) ISSN-L 1455-1616 ISSN 1456-954X ISSN 1455-1616 http://tampub.uta.fi Suomen Yliopistopaino Oy – Juvenes Print Tampere 2018 441 729 Painotuote CONTENTS List of original communications ................................................................................................ -

Amphioxus Adaptive Immune System: the Insights from Genes
The Journal of Immunology Genes “Waiting” for Recruitment by the Adaptive Immune System: The Insights from Amphioxus1 Cuiling Yu,2* Meiling Dong,2* Xiaokun Wu,2* Shengguo Li,§ Shengfeng Huang,* Jing Su,* Jianwen Wei,* Yang Shen,* Chunyan Mou,* Xiaojin Xie,* Jianghai Lin,* Shaochun Yuan,* Xuesong Yu,* Yanhong Yu,* Jingchun Du,* Shicui Zhang,† Xuanxian Peng,‡ Mengqing Xiang,§ and Anlong Xu3* In seeking evidence of the existence of adaptive immune system (AIS) in ancient chordate, cDNA clones of six libraries from a protochordate, the Chinese amphioxus, were sequenced. Although the key molecules such as TCR, MHC, Ig, and RAG in AIS have not been identified from our database, we demonstrated in this study the extensive molecular evidence for the presence of genes homologous to many genes that are involved in AIS directly or indirectly, including some of which may represent the putative precursors of vertebrate AIS-related genes. The comparative analyses of these genes in different model organisms revealed the different fates of these genes during evolution. Their gene expression pattern suggested that the primitive digestive system is the pivotal place of the origin and evolution of the AIS. Our studies support the general statement that AIS appears after the jawless/jawed vertebrate split. However our study further reveals the fact that AIS is in its twilight in amphioxus and the evolution of the molecules in amphioxus are waiting for recruitment by the emergence of AIS. The Journal of Immunology, 2005, 174: 3493–3500. he hallmark of the adaptive immune system (AIS)4 is the brate (2, 3). The studies for the origin of the AIS focus on many presence of cells and molecules participating in the im- aspects: the origin of the Ag receptor, Ag processing and presen- T mune recognition of foreign pathogens and the memory tation system, and the effector cells (3, 4). -

The UVB-Induced Gene Expression Profile of Human Epidermis in Vivo Is Different from That of Cultured Keratinocytes
Oncogene (2006) 25, 2601–2614 & 2006 Nature Publishing Group All rights reserved 0950-9232/06 $30.00 www.nature.com/onc ORIGINAL ARTICLE The UVB-induced gene expression profile of human epidermis in vivo is different from that of cultured keratinocytes CD Enk1, J Jacob-Hirsch2, H Gal3, I Verbovetski4, N Amariglio2, D Mevorach4, A Ingber1, D Givol3, G Rechavi2 and M Hochberg1 1Department of Dermatology, The Hadassah-Hebrew University Medical Center, Jerusalem, Israel; 2Department of Pediatric Hemato-Oncology and Functional Genomics, Safra Children’s Hospital, Sheba Medical Center and Sackler School of Medicine, Tel-Aviv University,Tel Aviv, Israel; 3Department of Molecular Cell Biology, Weizmann Institute of Science, Rehovot, Israel and 4The Laboratory for Cellular and Molecular Immunology, Department of Medicine, The Hadassah-Hebrew University Medical Center, Jerusalem, Israel In order to obtain a comprehensive picture of the radiation. UVB, with a wavelength range between 290 molecular events regulating cutaneous photodamage of and 320 nm, represents one of the most important intact human epidermis, suction blister roofs obtained environmental hazards affectinghuman skin (Hahn after a single dose of in vivo ultraviolet (UV)B exposure and Weinberg, 2002). To protect itself against the were used for microarray profiling. We found a changed DNA-damaging effects of sunlight, the skin disposes expression of 619 genes. Half of the UVB-regulated genes over highly complicated cellular programs, including had returned to pre-exposure baseline levels at 72 h, cell-cycle arrest, DNA repair and apoptosis (Brash et al., underscoring the transient character of the molecular 1996). Failure in selected elements of these defensive cutaneous UVB response. -

Diversification of Importin-Α Isoforms in Cellular Trafficking and Disease States
Thomas Jefferson University Jefferson Digital Commons Department of Biochemistry and Molecular Biology Department of Biochemistry and Molecular Biology Faculty Papers 2-15-2015 Diversification of importin-α isoforms in cellular trafficking and disease states. Ruth A. Pumroy Thomas Jefferson University, [email protected] Gino Cingolani Thomas Jefferson University, [email protected] Let us know how access to this document benefits ouy Follow this and additional works at: https://jdc.jefferson.edu/bmpfp Part of the Medical Biochemistry Commons Recommended Citation Pumroy, Ruth A. and Cingolani, Gino, "Diversification of importin-α isoforms in cellular trafficking and disease states." (2015). Department of Biochemistry and Molecular Biology Faculty Papers. Paper 107. https://jdc.jefferson.edu/bmpfp/107 This Article is brought to you for free and open access by the Jefferson Digital Commons. The effeJ rson Digital Commons is a service of Thomas Jefferson University's Center for Teaching and Learning (CTL). The ommonC s is a showcase for Jefferson books and journals, peer-reviewed scholarly publications, unique historical collections from the University archives, and teaching tools. The effeJ rson Digital Commons allows researchers and interested readers anywhere in the world to learn about and keep up to date with Jefferson scholarship. This article has been accepted for inclusion in Department of Biochemistry and Molecular Biology Faculty Papers by an authorized administrator of the Jefferson Digital Commons. For more information, please contact: [email protected]. HHS Public Access Author manuscript Author Manuscript Author ManuscriptBiochem Author Manuscript J. Author manuscript; Author Manuscript available in PMC 2015 April 21. Published in final edited form as: Biochem J. -

Stem Cells® Original Article
® Stem Cells Original Article Properties of Pluripotent Human Embryonic Stem Cells BG01 and BG02 XIANMIN ZENG,a TAKUMI MIURA,b YONGQUAN LUO,b BHASKAR BHATTACHARYA,c BRIAN CONDIE,d JIA CHEN,a IRENE GINIS,b IAN LYONS,d JOSEF MEJIDO,c RAJ K. PURI,c MAHENDRA S. RAO,b WILLIAM J. FREEDa aCellular Neurobiology Research Branch, National Institute on Drug Abuse, Department of Health and Human Services (DHHS), Baltimore, Maryland, USA; bLaboratory of Neuroscience, National Institute of Aging, DHHS, Baltimore, Maryland, USA; cLaboratory of Molecular Tumor Biology, Division of Cellular and Gene Therapies, Center for Biologics Evaluation and Research, Food and Drug Administration, Bethesda, Maryland, USA; dBresaGen Inc., Athens, Georgia, USA Key Words. Embryonic stem cells · Differentiation · Microarray ABSTRACT Human ES (hES) cell lines have only recently been compared with pooled human RNA. Ninety-two of these generated, and differences between human and mouse genes were also highly expressed in four other hES lines ES cells have been identified. In this manuscript we (TE05, GE01, GE09, and pooled samples derived from describe the properties of two human ES cell lines, GE01, GE09, and GE07). Included in the list are genes BG01 and BG02. By immunocytochemistry and reverse involved in cell signaling and development, metabolism, transcription polymerase chain reaction, undifferenti- transcription regulation, and many hypothetical pro- ated cells expressed markers that are characteristic of teins. Two focused arrays designed to examine tran- ES cells, including SSEA-3, SSEA-4, TRA-1-60, TRA-1- scripts associated with stem cells and with the 81, and OCT-3/4. Both cell lines were readily main- transforming growth factor-β superfamily were tained in an undifferentiated state and could employed to examine differentially expressed genes. -

Cytotoxicity, Anti-Angiogenic, Apoptotic Effects and Transcript Profiling of A
Kuete et al. Cell Division 2012, 7:16 http://www.celldiv.com/content/7/1/16 RESEARCH Open Access Cytotoxicity, anti-angiogenic, apoptotic effects and transcript profiling of a naturally occurring naphthyl butenone, guieranone A Victor Kuete1,2*, Tolga Eichhorn2, Benjamin Wiench2, Benjamin Krusche2 and Thomas Efferth2* Abstract Background: Malignant diseases are responsible of approximately 13% of all deaths each year in the world. Natural products represent a valuable source for the development of novel anticancer drugs. The present study was aimed at evaluating the cytotoxicity of a naphtyl butanone isolated from the leaves of Guiera senegalensis, guieranone A (GA). Results: The results indicated that GA was active on 91.67% of the 12 tested cancer cell lines, the IC50 values below 4 μg/ml being recorded on 83.33% of them. In addition, the IC50 values obtained on human lymphoblastic leukemia CCRF-CEM (0.73 μg/ml) and its resistant subline CEM/ADR5000 (1.01 μg/ml) and on lung adenocarcinoma A549 (0.72 μg/ml) cell lines were closer or lower than that of doxorubicin. Interestingly, low cytotoxicity to normal hepatocyte, AML12 cell line was observed. GA showed anti-angiogenic activity with up to 51.9% inhibition of the growth of blood capillaries on the chorioallantoic membrane of quail embryo. Its also induced apotosis and cell cycle arrest. Ingenuity Pathway Analysis identified several pathways in CCRF-CEM cells and functional group of genes regulated upon GA treatment (P < 0.05), the Cell Cycle: G2/M DNA Damage Checkpoint Regulation and ATM Signaling pathways being amongst the four most involved functional groups. -

(KPNA2) and Its Potential Novel Cargo Proteins in Nonsmall Cell Lung Cancer *□S
Research © 2012 by The American Society for Biochemistry and Molecular Biology, Inc. This paper is available on line at http://www.mcponline.org Quantitative Proteomics Reveals Regulation of Karyopherin Subunit Alpha-2 (KPNA2) and Its Potential Novel Cargo Proteins in Nonsmall Cell Lung Cancer *□S Chun-I Wang‡‡‡, Kun-Yi Chien‡¶, Chih-Liang Wangʈ, Hao-Ping Liu¶, Chia-Chen Cheng§, Yu-Sun Chang‡¶, Jau-Song Yu‡§¶, and Chia-Jung Yu‡§¶**‡‡ The process of nucleocytoplasmic shuttling is mediated cancer. Molecular & Cellular Proteomics 11: 10.1074/ by karyopherins. Dysregulated expression of karyo- mcp.M111.016592, 1105–1122, 2012. pherins may trigger oncogenesis through aberrant distri- bution of cargo proteins. Karyopherin subunit alpha-2 (KPNA2) was previously identified as a potential bio- Transportation of proteins and RNAs into (import) and out of marker for nonsmall cell lung cancer by integration of the (export) the nucleus occurs through the nuclear pore complex cancer cell secretome and tissue transcriptome data sets. and is a vital event in eukaryotic cells. Nucleocytoplasmic Knockdown of KPNA2 suppressed the proliferation and shuttling of the large complex (Ͼ40 kDa) is mediated by an migration abilities of lung cancer cells. However, the pre- evolutionarily conserved family of transport factors, desig- cise molecular mechanisms underlying KPNA2 activity in cancer remain to be established. In the current study, we nated karyopherins (1). The family of karyopherins, including applied gene knockdown, subcellular fractionation, and importins and exportins, share limited sequence identity (15– stable isotope labeling by amino acids in cell culture- 25%) but adopt similar conformations. In human cells, at least based quantitative proteomic strategies to systematically 22 importin  and 6 importin ␣ proteins have been identified to analyze the KPNA2-regulating protein profiles in an ade- date (2, 3). -

IRF1 Negatively Regulates Oncogenic KPNA2 Expression Under Growth Stimulation and Hypoxia in Lung Cancer Cells
OncoTargets and Therapy Dovepress open access to scientific and medical research Open Access Full Text Article ORIGINAL RESEARCH IRF1 Negatively Regulates Oncogenic KPNA2 Expression Under Growth Stimulation and Hypoxia in Lung Cancer Cells This article was published in the following Dove Press journal: OncoTargets and Therapy Jie-Xin Huang1 Purpose: Karyopherin alpha 2 (KPNA2) has been reported as an oncogenic protein in Yi-Cheng Wu2 numerous human cancers and is currently considered a potential therapeutic target. However, Ya-Yun Cheng 1 the transcriptional regulation and physiological conditions underlying KPNA2 expression Chih-Liang Wang3,4,* remain unclear. The aim of the present study was to investigate the role and regulation of – Chia-Jung Yu 1,4 6,* interferon regulatory factor-1 (IRF1) in modulating KPNA2 expression in lung adenocarci- noma (ADC). 1 Graduate Institute of Biomedical Materials and methods: Bioinformatics tools and chromatin immunoprecipitation were Sciences, College of Medicine, Chang Gung University, Taoyuan, Taiwan; used to analyze the transcription factor (TF) binding sites in the KPNA2 promoter region. We 2Department of Thoracic Surgery, Chang searched for a potential role of IRF1 in non-small-cell lung cancer (NSCLC) using Gung Memorial Hospital, Taoyuan, Oncomine and Kaplan-Meier Plotter datasets. qRT-PCR was applied to examine the role of Taiwan; 3School of Medicine, College of Medicine, Chang Gung University, IRF1 and signaling involved in regulating KPNA2 transcription. Western blotting was used Taoyuan, Taiwan; 4Division of Pulmonary to determine the effects of extracellular stimulation and intracellular signaling on the Oncology and Interventional Bronchoscopy, Department of Thoracic modulation of KPNA2-related TF expression. Medicine, Chang Gung Memorial Results: IRF1 was identified as a novel TF that suppresses KPNA2 gene expression. -

Karyopherin Alpha 1 Regulates Satellite Cell Proliferation And
Karyopherin Alpha 1 Regulates Satellite Cell Proliferation and Survival By Modulating Nuclear Import Hyojung Choo, Emory University Alicia Cutler, Emory University Franziska Rother, Max-Delbrück-Center for Molecular Medicine Michael Bader, Max-Delbrück-Center for Molecular Medicine Grace Pavlath, Emory University Journal Title: STEM CELLS Volume: Volume 34, Number 11 Publisher: AlphaMed Press | 2016-11-01, Pages 2784-2797 Type of Work: Article | Post-print: After Peer Review Publisher DOI: 10.1002/stem.2467 Permanent URL: https://pid.emory.edu/ark:/25593/s324r Final published version: http://dx.doi.org/10.1002/stem.2467 Copyright information: © 2016 AlphaMed Press Accessed September 23, 2021 10:27 AM EDT HHS Public Access Author manuscript Author ManuscriptAuthor Manuscript Author Stem Cells Manuscript Author . Author manuscript; Manuscript Author available in PMC 2017 June 01. Published in final edited form as: Stem Cells. 2016 November ; 34(11): 2784–2797. doi:10.1002/stem.2467. Karyopherin alpha 1 regulates satellite cell proliferation and survival by modulating nuclear import Hyo-Jung Choo1,*, Alicia Cutler1,2, Franziska Rother3, Michael Bader3, and Grace K. Pavlath1,* 1Department of Pharmacology, Emory University, Atlanta, GA 30322, USA 2Graduate Program in Biochemistry, Cell and Developmental Biology, Emory University, Atlanta, GA 30322, USA 3Max-Delbrück-Center for Molecular Medicine, Berlin-Buch, Germany Abstract Satellite cells are stem cells with an essential role in skeletal muscle repair. Precise regulation of gene expression is critical for proper satellite cell quiescence, proliferation, differentiation and self -renewal. Nuclear proteins required for gene expression are dependent on the nucleocytoplasmic transport machinery to access to nucleus, however little is known about regulation of nuclear transport in satellite cells. -
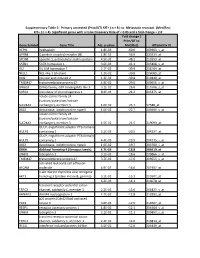
Gene Symbol Gene Title Adj. P-‐Value Fold Change ( Prim/UT to Met
Supplementary Table 2: Primary untreated (Prim/UT) KIT+ ( n = 8 ) vs. Metastatic resistant (Met/Res) KIT+ ( n = 4). Significant genes with a False Discovery Rate of < 0.05 and a fold change > 2.0 Fold change ( Prim/UT to Gene Symbol Gene Title Adj. p-value Met/Res) Affymetrix ID HEPH hephaestin 1.8E-03 -60.6 203903_s_at GPR88 G protein-coupled receptor 88 1.8E-02 -56.0 220313_at SPON1 spondin 1, extracellular matrix protein 4.5E-02 -48.2 213993_at SATB1 SATB homeobox 1 3.6E-03 -41.1 203408_s_at ISL1 ISL LIM homeobox 1 2.7E-02 -39.9 206104_at NELL1 NEL-like 1 (chicken) 1.2E-02 -39.8 206089_at RAI2 retinoic acid induced 2 1.3E-02 -30.8 219440_at TMEM47 transmembrane protein 47 4.9E-02 -29.0 209656_s_at DIRAS3 DIRAS family, GTP-binding RAS-like 3 3.1E-02 -26.8 215506_s_at SCRG1 stimulator of chondrogenesis 1 8.3E-03 -24.2 205475_at solute carrier family 24 (sodium/potassium/calcium SLC24A3 exchanger), member 3 1.6E-02 -23.7 57588_at DIO2 deiodinase, iodothyronine, type II 1.0E-02 -22.7 203699_s_at solute carrier family 24 (sodium/potassium/calcium SLC24A3 exchanger), member 3 1.5E-02 -21.5 219090_at GULP, engulfment adaptor PTB domain GULP1 containing 1 5.2E-03 -20.5 204237_at GULP, engulfment adaptor PTB domain GULP1 containing 1 4.4E-03 -19.9 204235_s_at DIO2 deiodinase, iodothyronine, type II 1.0E-02 -19.7 203700_s_at DKK4 dickkopf homolog 4 (Xenopus laevis) 4.7E-03 -18.8 206619_at LPHN3 latrophilin 3 3.1E-02 -18.6 209866_s_at TMEM47 transmembrane protein 47 3.7E-02 -17.6 209655_s_at activated leukocyte cell adhesion ALCAM molecule 4.9E-02 -

Importin Α2 Associates with Chromatin Via a Novel DNA Binding Domain
bioRxiv preprint doi: https://doi.org/10.1101/2020.05.04.075580; this version posted May 5, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Importin α2 associates with chromatin via a novel DNA binding domain Kazuya Jibiki1, Takashi Kodama2, 5, Atsushi Suenaga1,3, Yota Kawase1, Noriko Shibazaki3, Shin Nomoto1, Seiya Nagasawa3, Misaki Nagashima3, Shieri Shimodan3, Renan Kikuchi3, Noriko Saitoh4, Yoshihiro Yoneda5, Ken-ich Akagi5, 6, Noriko Yasuhara1, 3* 1 Graduate School of Integrated Basic Sciences, Nihon University, Setagaya-ku, Tokyo, Japan 2 Laboratory of Molecular Biophysics, Institute for Protein Research, Osaka University, Osaka, Japan 3 Department of Biosciences, College of Humanities and Sciences, Nihon University, Setagaya- ku, Tokyo, Japan 4 Division of Cancer Biology, The Cancer Institute of JFCR, Tokyo, Japan 5 National Institute of Biomedical Innovation, Health and Nutrition (NIBIOHN), Osaka, Japan 6 present address: Environmental Metabolic Analysis Research Team, RIKEN Center for Sustainable Resource Science, Yokohama, Kanagawa, Japan * Corresponding author. E-mail: [email protected] bioRxiv preprint doi: https://doi.org/10.1101/2020.05.04.075580; this version posted May 5, 2020. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Abstract The nuclear transport of functional proteins is important for facilitating appropriate gene expression. The proteins of the importin α family of nuclear transport receptors operate via several pathways to perform their nuclear protein import function. Additionally, these proteins are also reported to possess other functions, including chromatin association and gene regulation. -

A GENOMIC SCREEN for ZIC1 TARGET GENES in NEURAL DEVELOPMENT by Shuzhao Li a Thesis Submitted in Partial Fulfillment of the Requ
A GENOMIC SCREEN FOR ZIC1 TARGET GENES IN NEURAL DEVELOPMENT by Shuzhao Li A thesis submitted in partial fulfillment of the requirements for the degree of Master of Science in Biological Sciences MONTANA STATE UNIVERSITY Bozeman, Montana July 2006 ©COPYRIGHT by Shuzhao Li 2006 All Rights Reserved ii APPROVAL of a thesis submitted by Shuzhao Li This thesis has been read by each member of the thesis committee and has been found to be satisfactory regarding content, English usage, format, citations, bibliographic style, and consistency, and is ready for submission to the Division of Graduate Education. Dr. Christa Merzdorf Approved for the Department of Cell Biology and Neuroscience Dr. Gwen Jacobs Approved for the Division of Graduate Education Dr. Joseph Fedock iii STATEMENT OF PERMISSION TO USE In presenting this thesis in partial fulfillment of the requirements for a master’s degree at Montana State University, I agree that the Library shall make it available to borrowers under rules of the Library. If I have indicated my intention to copyright this thesis by including a copyright notice page, copying is allowable only for scholarly purposes, consistent with “fair use” as prescribed in the U.S. Copyright Law. Requests for permission for extended quotation from or reproduction of this thesis in whole or in parts may be granted only by the copyright holder. Shuzhao Li July 2006 iv TABLE OF CONTENTS 1. INTRODUCTION.......................................................................................................1 2. GENOMIC SCREEN WITH