In Silico Identification and Comparative Analysis of Differentially Expressed Genes in Human and Mouse Tissues Sheng-Ying Pao1,2, Win-Li Lin1 and Ming-Jing Hwang*2
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Aneuploidy: Using Genetic Instability to Preserve a Haploid Genome?
Health Science Campus FINAL APPROVAL OF DISSERTATION Doctor of Philosophy in Biomedical Science (Cancer Biology) Aneuploidy: Using genetic instability to preserve a haploid genome? Submitted by: Ramona Ramdath In partial fulfillment of the requirements for the degree of Doctor of Philosophy in Biomedical Science Examination Committee Signature/Date Major Advisor: David Allison, M.D., Ph.D. Academic James Trempe, Ph.D. Advisory Committee: David Giovanucci, Ph.D. Randall Ruch, Ph.D. Ronald Mellgren, Ph.D. Senior Associate Dean College of Graduate Studies Michael S. Bisesi, Ph.D. Date of Defense: April 10, 2009 Aneuploidy: Using genetic instability to preserve a haploid genome? Ramona Ramdath University of Toledo, Health Science Campus 2009 Dedication I dedicate this dissertation to my grandfather who died of lung cancer two years ago, but who always instilled in us the value and importance of education. And to my mom and sister, both of whom have been pillars of support and stimulating conversations. To my sister, Rehanna, especially- I hope this inspires you to achieve all that you want to in life, academically and otherwise. ii Acknowledgements As we go through these academic journeys, there are so many along the way that make an impact not only on our work, but on our lives as well, and I would like to say a heartfelt thank you to all of those people: My Committee members- Dr. James Trempe, Dr. David Giovanucchi, Dr. Ronald Mellgren and Dr. Randall Ruch for their guidance, suggestions, support and confidence in me. My major advisor- Dr. David Allison, for his constructive criticism and positive reinforcement. -

The Conserved DNMT1-Dependent Methylation Regions in Human Cells
Freeman et al. Epigenetics & Chromatin (2020) 13:17 https://doi.org/10.1186/s13072-020-00338-8 Epigenetics & Chromatin RESEARCH Open Access The conserved DNMT1-dependent methylation regions in human cells are vulnerable to neurotoxicant rotenone exposure Dana M. Freeman1 , Dan Lou1, Yanqiang Li1, Suzanne N. Martos1 and Zhibin Wang1,2,3* Abstract Background: Allele-specifc DNA methylation (ASM) describes genomic loci that maintain CpG methylation at only one inherited allele rather than having coordinated methylation across both alleles. The most prominent of these regions are germline ASMs (gASMs) that control the expression of imprinted genes in a parent of origin-dependent manner and are associated with disease. However, our recent report reveals numerous ASMs at non-imprinted genes. These non-germline ASMs are dependent on DNA methyltransferase 1 (DNMT1) and strikingly show the feature of random, switchable monoallelic methylation patterns in the mouse genome. The signifcance of these ASMs to human health has not been explored. Due to their shared allelicity with gASMs, herein, we propose that non-tradi- tional ASMs are sensitive to exposures in association with human disease. Results: We frst explore their conservancy in the human genome. Our data show that our putative non-germline ASMs were in conserved regions of the human genome and located adjacent to genes vital for neuronal develop- ment and maturation. We next tested the hypothesized vulnerability of these regions by exposing human embryonic kidney cell HEK293 with the neurotoxicant rotenone for 24 h. Indeed,14 genes adjacent to our identifed regions were diferentially expressed from RNA-sequencing. We analyzed the base-resolution methylation patterns of the predicted non-germline ASMs at two neurological genes, HCN2 and NEFM, with potential to increase the risk of neurodegenera- tion. -

Characterization of the Macrophage Transcriptome in Glomerulonephritis-Susceptible and -Resistant Rat Strains
Genes and Immunity (2011) 12, 78–89 & 2011 Macmillan Publishers Limited All rights reserved 1466-4879/11 www.nature.com/gene ORIGINAL ARTICLE Characterization of the macrophage transcriptome in glomerulonephritis-susceptible and -resistant rat strains K Maratou1, J Behmoaras2, C Fewings1, P Srivastava1, Z D’Souza1, J Smith3, L Game4, T Cook2 and T Aitman1 1Physiological Genomics and Medicine Group, MRC Clinical Sciences Centre, Imperial College London, London, UK; 2Centre for Complement and Inflammation Research, Imperial College London, London, UK; 3Renal Section, Imperial College London, London, UK and 4Genomics Laboratory, MRC Clinical Sciences Centre, London, UK Crescentic glomerulonephritis (CRGN) is a major cause of rapidly progressive renal failure for which the underlying genetic basis is unknown. Wistar–Kyoto (WKY) rats show marked susceptibility to CRGN, whereas Lewis rats are resistant. Glomerular injury and crescent formation are macrophage dependent and mainly explained by seven quantitative trait loci (Crgn1–7). Here, we used microarray analysis in basal and lipopolysaccharide (LPS)-stimulated macrophages to identify genes that reside on pathways predisposing WKY rats to CRGN. We detected 97 novel positional candidates for the uncharacterized Crgn3–7. We identified 10 additional secondary effector genes with profound differences in expression between the two strains (45-fold change, o1% false discovery rate) for basal and LPS-stimulated macrophages. Moreover, we identified eight genes with differentially expressed alternatively spliced isoforms, by using an in-depth analysis at the probe level that allowed us to discard false positives owing to polymorphisms between the two rat strains. Pathway analysis identified several common linked pathways, enriched for differentially expressed genes, which affect macrophage activation. -
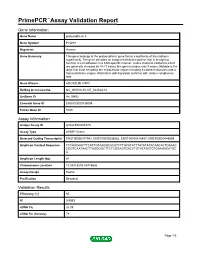
Primepcr™Assay Validation Report
PrimePCR™Assay Validation Report Gene Information Gene Name protocadherin 8 Gene Symbol PCDH8 Organism Human Gene Summary This gene belongs to the protocadherin gene family a subfamily of the cadherin superfamily. The gene encodes an integral membrane protein that is thought to function in cell adhesion in a CNS-specific manner. Unlike classical cadherins which are generally encoded by 15-17 exons this gene includes only 3 exons. Notable is the large first exon encoding the extracellular region including 6 cadherin domains and a transmembrane region. Alternative splicing yields isoforms with unique cytoplasmic tails. Gene Aliases ARCADLIN, PAPC RefSeq Accession No. NC_000013.10, NT_024524.14 UniGene ID Hs.19492 Ensembl Gene ID ENSG00000136099 Entrez Gene ID 5100 Assay Information Unique Assay ID qHsaCED0006870 Assay Type SYBR® Green Detected Coding Transcript(s) ENST00000377942, ENST00000338862, ENST00000418407, ENST00000448969 Amplicon Context Sequence TCCAACAACTTCATTGTAAGGCACATCTTGCATATTTATATATACAACACTGAAAC CGGTCAATAACTTAGGGGCTTCTCGGAGTGACCTGTATATGTGTGAAGACATGC A Amplicon Length (bp) 81 Chromosome Location 13:53418576-53418686 Assay Design Exonic Purification Desalted Validation Results Efficiency (%) 95 R2 0.9993 cDNA Cq 26.09 cDNA Tm (Celsius) 78 Page 1/5 PrimePCR™Assay Validation Report gDNA Cq 25.07 Specificity (%) 100 Information to assist with data interpretation is provided at the end of this report. Page 2/5 PrimePCR™Assay Validation Report PCDH8, Human Amplification Plot Amplification of cDNA generated from 25 ng of universal -

Nº Ref Uniprot Proteína Péptidos Identificados Por MS/MS 1 P01024
Document downloaded from http://www.elsevier.es, day 26/09/2021. This copy is for personal use. Any transmission of this document by any media or format is strictly prohibited. Nº Ref Uniprot Proteína Péptidos identificados 1 P01024 CO3_HUMAN Complement C3 OS=Homo sapiens GN=C3 PE=1 SV=2 por 162MS/MS 2 P02751 FINC_HUMAN Fibronectin OS=Homo sapiens GN=FN1 PE=1 SV=4 131 3 P01023 A2MG_HUMAN Alpha-2-macroglobulin OS=Homo sapiens GN=A2M PE=1 SV=3 128 4 P0C0L4 CO4A_HUMAN Complement C4-A OS=Homo sapiens GN=C4A PE=1 SV=1 95 5 P04275 VWF_HUMAN von Willebrand factor OS=Homo sapiens GN=VWF PE=1 SV=4 81 6 P02675 FIBB_HUMAN Fibrinogen beta chain OS=Homo sapiens GN=FGB PE=1 SV=2 78 7 P01031 CO5_HUMAN Complement C5 OS=Homo sapiens GN=C5 PE=1 SV=4 66 8 P02768 ALBU_HUMAN Serum albumin OS=Homo sapiens GN=ALB PE=1 SV=2 66 9 P00450 CERU_HUMAN Ceruloplasmin OS=Homo sapiens GN=CP PE=1 SV=1 64 10 P02671 FIBA_HUMAN Fibrinogen alpha chain OS=Homo sapiens GN=FGA PE=1 SV=2 58 11 P08603 CFAH_HUMAN Complement factor H OS=Homo sapiens GN=CFH PE=1 SV=4 56 12 P02787 TRFE_HUMAN Serotransferrin OS=Homo sapiens GN=TF PE=1 SV=3 54 13 P00747 PLMN_HUMAN Plasminogen OS=Homo sapiens GN=PLG PE=1 SV=2 48 14 P02679 FIBG_HUMAN Fibrinogen gamma chain OS=Homo sapiens GN=FGG PE=1 SV=3 47 15 P01871 IGHM_HUMAN Ig mu chain C region OS=Homo sapiens GN=IGHM PE=1 SV=3 41 16 P04003 C4BPA_HUMAN C4b-binding protein alpha chain OS=Homo sapiens GN=C4BPA PE=1 SV=2 37 17 Q9Y6R7 FCGBP_HUMAN IgGFc-binding protein OS=Homo sapiens GN=FCGBP PE=1 SV=3 30 18 O43866 CD5L_HUMAN CD5 antigen-like OS=Homo -

Identification of DNA Methylation Associated Gene Signatures in Endometrial Cancer Via Integrated Analysis of DNA Methylation and Gene Expression Systematically
J Gynecol Oncol. 2017 Nov;28(6):e83 https://doi.org/10.3802/jgo.2017.28.e83 pISSN 2005-0380·eISSN 2005-0399 Original Article Identification of DNA methylation associated gene signatures in endometrial cancer via integrated analysis of DNA methylation and gene expression systematically Chuandi Men ,1,2 Hongjuan Chai ,1 Xumin Song ,1 Yue Li ,1 Huawen Du ,1 Qing Ren 1 1Department of Gynecology and Obstetrics, Shanghai Ninth People's Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China Received: May 11, 2017 2Graduate School, Bengbu Medical College, Bengbu, China Revised: Aug 2, 2017 Accepted: Aug 10, 2017 Correspondence to ABSTRACT Qing Ren Department of Gynecology and Obstetrics, Objective: Endometrial cancer (EC) is a common gynecologic cancer worldwide. However, Shanghai Ninth People's Hospital, Shanghai the pathogenesis of EC has not been epigenetically elucidated. Here, this study aims Jiao Tong University School of Medicine, No. to describe the DNA methylation profile and identify favorable gene signatures highly 280, Mohe Road, Baoshan District, Shanghai 201900, China. associated with aberrant DNA methylation changes in EC. E-mail: [email protected] Methods: The data regarding DNA methylation and gene expression were downloaded from The Cancer Genome Atlas (TCGA) database. Differentially methylated CpG sites (DMCs), Copyright © 2017. Asian Society of differentially methylated regions (DMRs), and differentially expressed genes (DEGs) were Gynecologic Oncology, Korean Society of Gynecologic Oncology identified, and the relationship between the 2 omics was further analyzed. In addition, This is an Open Access article distributed weighted CpG site co-methylation network (WCCN) was constructed followed by an under the terms of the Creative Commons integrated analysis of DNA methylation and gene expression data. -

The GALNT9, BNC1 and CCDC8 Genes Are Frequently Epigenetically Dysregulated in Breast Tumours That Metastasise to the Brain Rajendra P
Pangeni et al. Clinical Epigenetics (2015) 7:57 DOI 10.1186/s13148-015-0089-x RESEARCH Open Access The GALNT9, BNC1 and CCDC8 genes are frequently epigenetically dysregulated in breast tumours that metastasise to the brain Rajendra P. Pangeni1, Prasanna Channathodiyil1, David S. Huen2, Lawrence W. Eagles1, Balraj K. Johal2, Dawar Pasha2, Natasa Hadjistephanou2, Oliver Nevell2, Claire L. Davies2, Ayobami I. Adewumi2, Hamida Khanom2, Ikroop S. Samra2, Vanessa C. Buzatto2, Preethi Chandrasekaran2, Thoraia Shinawi3, Timothy P. Dawson4, Katherine M. Ashton4, Charles Davis4, Andrew R. Brodbelt5, Michael D. Jenkinson5, Ivan Bièche6, Farida Latif3, John L. Darling1, Tracy J. Warr1 and Mark R. Morris1,2,3* Abstract Background: Tumour metastasis to the brain is a common and deadly development in certain cancers; 18–30 % of breast tumours metastasise to the brain. The contribution that gene silencing through epigenetic mechanisms plays in these metastatic tumours is not well understood. Results: We have carried out a bioinformatic screen of genome-wide breast tumour methylation data available at The Cancer Genome Atlas (TCGA) and a broad literature review to identify candidate genes that may contribute to breast to brain metastasis (BBM). This analysis identified 82 candidates. We investigated the methylation status of these genes using Combined Bisulfite and Restriction Analysis (CoBRA) and identified 21 genes frequently methylated in BBM. We have identified three genes, GALNT9, CCDC8 and BNC1, that were frequently methylated (55, 73 and 71 %, respectively) and silenced in BBM and infrequently methylated in primary breast tumours. CCDC8 was commonly methylated in brain metastases and their associated primary tumours whereas GALNT9 and BNC1 were methylated and silenced only in brain metastases, but not in the associated primary breast tumours from individual patients. -

Genotype-Phenotype Correlations in Patients with Retinoblastoma And
Genotype-phenotype correlations in patients with Retinoblastoma and an interstitial 13q deletion Diana Mitter, Reinhard Ullmann, Artur Muradyan, Ludger Klein-Hitpaß, Deniz Kanber, Dietmar R Lohmann, Katrin Ounap, Marc Kaulisch To cite this version: Diana Mitter, Reinhard Ullmann, Artur Muradyan, Ludger Klein-Hitpaß, Deniz Kanber, et al.. Genotype-phenotype correlations in patients with Retinoblastoma and an interstitial 13q deletion. European Journal of Human Genetics, Nature Publishing Group, 2011, 10.1038/ejhg.2011.58. hal- 00633984 HAL Id: hal-00633984 https://hal.archives-ouvertes.fr/hal-00633984 Submitted on 20 Oct 2011 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. 1 Genotype-phenotype correlations in patients with Retinoblastoma and interstitial 13q deletions 1, *Diana Mitter, 2Reinhard Ullmann, 2Artur Muradyan, 3Ludger Klein-Hitpaß, 1Deniz Kanber, 4Katrin Õunap, 5Marc Kaulisch, 1Dietmar Lohmann 1Institut für Humangenetik, Universitätsklinikum Essen, Germany; 2Max-Planck- Institute of Molecular Genetic, Berlin, Germany; 3Institute for Cell Biology (Tumor Research), University of Duisburg-Essen, Germany; 4Department of Genetics, United Laboratories, Tartu University Hospital, Estonia; 5Institute for Research Information and Quality Assurance, Germany. *Correspondence and present address: Diana Mitter, Institut für Humangenetik, Universitätsklinikum Leipzig, Philipp-Rosenthal-Str. -

Bioinformatics Tools and Applications for Rainbow Trout by Rafet Al
Bioinformatics Tools and Applications for Rainbow Trout By Rafet Al-Tobasei A Dissertation Submitted in Partial Fulfillment Of the Requirements for Degree of Doctor of Philosophy in Computational Science Middle Tennessee State University May 2017 Dissertation Committee: Dr. Mohamed Salem (Chair) Dr. Hyrum D. Carroll Dr. Cen Li Dr. Rebecca Seipelt -Thiemann Dr. Joshua L. Phillips Dedicated to My children (Ahmad, Zayna, Ruba, Leena), my lovely wife (Nour Kattih), and my Mother (Sadeqah Alkatib) ii ACKNOWLEDGMENTS During my study, I was blessed with many people that supported me and encouraged me to make this PhD a reality. My Advisor, Dr. Mohamed Salem, I do not know how to thank him enough for his support and guidance through this journey. Dr. Salem went far and beyond what a dissertation chair does. He taught me about a subject that was completely out of my radar. He helped me drafting my research with patience and excellent enlightenment. Working with Dr. Salem has been a pleasure that will influence me forever. Computational Science Director, Dr. John Wallin, thanks for all the supports and funds you provided me. Your support and encouragements will always be appreciated. I would like to thank Dr. Chrisila Pettey, Computer Science department chair, for her support. I always enjoy working with Dr. Hyrum Carroll sharing ideas and thoughts. Dr. Cen Li; I always enjoyed your classes as a student and as a lab assistant. I learned from you how to be a better instructor. I would like to thank my doctoral committee members: Dr. Rebecca Seipelt - Thiemann and Dr. Joshua Philips for their support, time, feedback, and assistance reviewing this dissertation. -

CREB-Dependent Transcription in Astrocytes: Signalling Pathways, Gene Profiles and Neuroprotective Role in Brain Injury
CREB-dependent transcription in astrocytes: signalling pathways, gene profiles and neuroprotective role in brain injury. Tesis doctoral Luis Pardo Fernández Bellaterra, Septiembre 2015 Instituto de Neurociencias Departamento de Bioquímica i Biologia Molecular Unidad de Bioquímica y Biologia Molecular Facultad de Medicina CREB-dependent transcription in astrocytes: signalling pathways, gene profiles and neuroprotective role in brain injury. Memoria del trabajo experimental para optar al grado de doctor, correspondiente al Programa de Doctorado en Neurociencias del Instituto de Neurociencias de la Universidad Autónoma de Barcelona, llevado a cabo por Luis Pardo Fernández bajo la dirección de la Dra. Elena Galea Rodríguez de Velasco y la Dra. Roser Masgrau Juanola, en el Instituto de Neurociencias de la Universidad Autónoma de Barcelona. Doctorando Directoras de tesis Luis Pardo Fernández Dra. Elena Galea Dra. Roser Masgrau In memoriam María Dolores Álvarez Durán Abuela, eres la culpable de que haya decidido recorrer el camino de la ciencia. Que estas líneas ayuden a conservar tu recuerdo. A mis padres y hermanos, A Meri INDEX I Summary 1 II Introduction 3 1 Astrocytes: physiology and pathology 5 1.1 Anatomical organization 6 1.2 Origins and heterogeneity 6 1.3 Astrocyte functions 8 1.3.1 Developmental functions 8 1.3.2 Neurovascular functions 9 1.3.3 Metabolic support 11 1.3.4 Homeostatic functions 13 1.3.5 Antioxidant functions 15 1.3.6 Signalling functions 15 1.4 Astrocytes in brain pathology 20 1.5 Reactive astrogliosis 22 2 The transcription -

Robles JTO Supplemental Digital Content 1
Supplementary Materials An Integrated Prognostic Classifier for Stage I Lung Adenocarcinoma based on mRNA, microRNA and DNA Methylation Biomarkers Ana I. Robles1, Eri Arai2, Ewy A. Mathé1, Hirokazu Okayama1, Aaron Schetter1, Derek Brown1, David Petersen3, Elise D. Bowman1, Rintaro Noro1, Judith A. Welsh1, Daniel C. Edelman3, Holly S. Stevenson3, Yonghong Wang3, Naoto Tsuchiya4, Takashi Kohno4, Vidar Skaug5, Steen Mollerup5, Aage Haugen5, Paul S. Meltzer3, Jun Yokota6, Yae Kanai2 and Curtis C. Harris1 Affiliations: 1Laboratory of Human Carcinogenesis, NCI-CCR, National Institutes of Health, Bethesda, MD 20892, USA. 2Division of Molecular Pathology, National Cancer Center Research Institute, Tokyo 104-0045, Japan. 3Genetics Branch, NCI-CCR, National Institutes of Health, Bethesda, MD 20892, USA. 4Division of Genome Biology, National Cancer Center Research Institute, Tokyo 104-0045, Japan. 5Department of Chemical and Biological Working Environment, National Institute of Occupational Health, NO-0033 Oslo, Norway. 6Genomics and Epigenomics of Cancer Prediction Program, Institute of Predictive and Personalized Medicine of Cancer (IMPPC), 08916 Badalona (Barcelona), Spain. List of Supplementary Materials Supplementary Materials and Methods Fig. S1. Hierarchical clustering of based on CpG sites differentially-methylated in Stage I ADC compared to non-tumor adjacent tissues. Fig. S2. Confirmatory pyrosequencing analysis of DNA methylation at the HOXA9 locus in Stage I ADC from a subset of the NCI microarray cohort. 1 Fig. S3. Methylation Beta-values for HOXA9 probe cg26521404 in Stage I ADC samples from Japan. Fig. S4. Kaplan-Meier analysis of HOXA9 promoter methylation in a published cohort of Stage I lung ADC (J Clin Oncol 2013;31(32):4140-7). Fig. S5. Kaplan-Meier analysis of a combined prognostic biomarker in Stage I lung ADC. -

13Q Deletion Syndrome Involving RB1: Characterization of a New Minimal Critical Region for Psychomotor Delay
G C A T T A C G G C A T genes Article 13q Deletion Syndrome Involving RB1: Characterization of a New Minimal Critical Region for Psychomotor Delay Flavia Privitera 1,2, Arianna Calonaci 3, Gabriella Doddato 1,2, Filomena Tiziana Papa 1,2, Margherita Baldassarri 1,2 , Anna Maria Pinto 4, Francesca Mari 1,2,4, Ilaria Longo 4, Mauro Caini 3, Daniela Galimberti 3, Theodora Hadjistilianou 5, Sonia De Francesco 5, Alessandra Renieri 1,2,4 and Francesca Ariani 1,2,4,* 1 Medical Genetics, University of Siena, 53100 Siena, Italy; fl[email protected] (F.P.); [email protected] (G.D.); fi[email protected] (F.T.P.); [email protected] (M.B.); [email protected] (F.M.); [email protected] (A.R.) 2 Med Biotech Hub and Competence Center, Department of Medical Biotechnologies, University of Siena, 53100 Siena, Italy 3 Unit of Pediatrics, Department of Maternal, Newborn and Child Health, Azienda Ospedaliera Universitaria Senese, Policlinico ‘Santa Maria alle Scotte’, 53100 Siena, Italy; [email protected] (A.C.); [email protected] (M.C.); [email protected] (D.G.) 4 Genetica Medica, Azienda Ospedaliera Universitaria Senese, 53100 Siena, Italy; [email protected] (A.M.P.); [email protected] (I.L.) 5 Unit of Ophthalmology and Retinoblastoma Referral Center, Department of Surgery, University of Siena, Policlinico ‘Santa Maria alle Scotte’, 53100 Siena, Italy; [email protected] (T.H.); [email protected] (S.D.F.) * Correspondence: [email protected]; Tel.: +39-057-723-3303 Citation: Privitera, F.; Calonaci, A.; Abstract: Retinoblastoma (RB) is an ocular tumor of the pediatric age caused by biallelic inactivation Doddato, G.; Papa, F.T.; Baldassarri, of the RB1 gene (13q14).