1 Psi News Letter January-2019
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Insights from a Pan India Sero- Epidemiological Survey
SHORT REPORT Insights from a Pan India Sero- Epidemiological survey (Phenome-India Cohort) for SARS-CoV2 Salwa Naushin1,2†, Viren Sardana1,2†, Rajat Ujjainiya1,2, Nitin Bhatheja1, Rintu Kutum1,2, Akash Kumar Bhaskar1,2, Shalini Pradhan1, Satyartha Prakash1, Raju Khan2,3, Birendra Singh Rawat2,4, Karthik Bharadwaj Tallapaka5, Mahesh Anumalla5, Giriraj Ratan Chandak2,5, Amit Lahiri2,6, Susanta Kar2,6, Shrikant Ramesh Mulay2,6, Madhav Nilakanth Mugale2,6, Mrigank Srivastava2,6, Shaziya Khan2,6, Anjali Srivastava2,6, Bhawana Tomar2,6, Murugan Veerapandian2,7, Ganesh Venkatachalam2,7, Selvamani Raja Vijayakumar7, Ajay Agarwal2,8, Dinesh Gupta8, Prakash M Halami2,9, Muthukumar Serva Peddha2,9, Gopinath M Sundaram2,9, Ravindra P Veeranna2,9, Anirban Pal2,10, Vinay Kumar Agarwal10, Anil Ku Maurya10, Ranvijay Kumar Singh2,11, Ashok Kumar Raman11, Suresh Kumar Anandasadagopan2,12, Parimala Karuppanan12, Subramanian Venkatesan2,12, Harish Kumar Sardana13, Anamika Kothari13, Rishabh Jain2,13, Anupama Thakur2,13, Devendra Singh Parihar2,13, Anas Saifi2,13, Jasleen Kaur2,13, Virendra Kumar13, Avinash Mishra2,14, Iranna Gogeri2,15, Geethavani Rayasam2,16, Praveen Singh1,2, Rahul Chakraborty1,2, Gaura Chaturvedi1,2, Pinreddy Karunakar1,2, Rohit Yadav1,2, Sunanda Singhmar1, Dayanidhi Singh1,2, Sharmistha Sarkar1,2, Purbasha Bhattacharya1,2, Sundaram Acharya1,2, Vandana Singh1,2, Shweta Verma1,2, Drishti Soni1,2, Surabhi Seth1,2, Sakshi Vashisht1,2, Sarita Thakran1,2, Firdaus Fatima1,2, Akash Pratap Singh1,2, Akanksha Sharma1,2, Babita Sharma1,2, *For -

Profile Prof. Gopal C. Kundu Has Obtained His Ph.D. in Chemistry
Profile Prof. Gopal C. Kundu has obtained his Ph.D. in Chemistry from Bose Institute, Kolkata, India (1989). He did his post-doctoral work at the Cleveland Clinic Foundation, University of Colorado, University of Wyoming, and the National Institutes of Health in USA in the area of cardiovascular research, inflammatory diseases and reproductive biology from 1989 to 1998. He joined as Scientist-D at National Centre for Cell Science (NCCS) in 1998 and then promoted to Scientist-E, - F and G. His area of research at NCCS is tumor biology, cancer stem cells, tumor- stroma interaction, angiogenesis, cancer therapeutics biomarker development and nanomedicine. He has received several awards including Fellows Award for Research Excellence from NIH, USA; National Bioscience Award from DBT, New Delhi; Shanti Swarup Bhatnagar Prize from CSIR, New Delhi; International Award in Oncology from Greece; International Young Investigator Award from Mayo Clinic, USA and 7th National Grassroots Innovation Award from Rashtrapati Bhavan, India. He is Fellow of National Academy of Sciences and Indian Academy of Sciences. He has published 93 papers including Nature Medicine, Science, PNAS (USA), Cancer Research, J. of Invest. Dermatology, J. of Biol. Chem, Trends in Cell Biol., Oncogene, Nanomedicine (London), Molecular Cancer, Nanoscale, Carcinogenesis, EMBO Reports, BMC Cancer etc and one US patent and two Indian Patents (Provisional). He serves as Editorial Board Member of Current Molecular Medicine, Current Chemical Biology, Molecular Medicine Reports, American Journal of Cancer Research and Associate Editor of Molecular Cancer and Journal of Cancer Metastasis and Treatment (JCMT). Prof. Kundu is an Adjunct Visiting Professor at Curtin University, Perth, Australia. -

List of Life Members As on 20Th January 2021
LIST OF LIFE MEMBERS AS ON 20TH JANUARY 2021 10. Dr. SAURABH CHANDRA SAXENA(2154) ALIGARH S/O NAGESH CHANDRA SAXENA POST HARDNAGANJ 1. Dr. SAAD TAYYAB DIST ALIGARH 202 125 UP INTERDISCIPLINARY BIOTECHNOLOGY [email protected] UNIT, ALIGARH MUSLIM UNIVERSITY ALIGARH 202 002 11. Dr. SHAGUFTA MOIN (1261) [email protected] DEPT. OF BIOCHEMISTRY J. N. MEDICAL COLLEGE 2. Dr. HAMMAD AHMAD SHADAB G. G.(1454) ALIGARH MUSLIM UNIVERSITY 31 SECTOR OF GENETICS ALIGARH 202 002 DEPT. OF ZOOLOGY ALIGARH MUSLIM UNIVERSITY 12. SHAIK NISAR ALI (3769) ALIGARH 202 002 DEPT. OF BIOCHEMISTRY FACULTY OF LIFE SCIENCE 3. Dr. INDU SAXENA (1838) ALIGARH MUSLIM UNIVERSITY, ALIGARH 202 002 HIG 30, ADA COLONY [email protected] AVANTEKA PHASE I RAMGHAT ROAD, ALIGARH 202 001 13. DR. MAHAMMAD REHAN AJMAL KHAN (4157) 4/570, Z-5, NOOR MANZIL COMPOUND 4. Dr. (MRS) KHUSHTAR ANWAR SALMAN(3332) DIDHPUR, CIVIL LINES DEPT. OF BIOCHEMISTRY ALIGARH UP 202 002 JAWAHARLAL NEHRU MEDICAL COLLEGE [email protected] ALIGARH MUSLIM UNIVERSITY ALIGARH 202 002 14. DR. HINA YOUNUS (4281) [email protected] INTERDISCIPLINARY BIOTECHNOLOGY UNIT ALIGARH MUSLIM UNIVERSITY 5. Dr. MOHAMMAD TABISH (2226) ALIGARH U.P. 202 002 DEPT. OF BIOCHEMISTRY [email protected] FACULTY OF LIFE SCIENCES ALIGARH MUSLIM UNIVERSITY 15. DR. IMTIYAZ YOUSUF (4355) ALIGARH 202 002 DEPT OF CHEMISTRY, [email protected] ALIGARH MUSLIM UNIVERSITY, ALIGARH, UP 202002 6. Dr. MOHAMMAD AFZAL (1101) [email protected] DEPT. OF ZOOLOGY [email protected] ALIGARH MUSLIM UNIVERSITY ALIGARH 202 002 ALLAHABAD 7. Dr. RIAZ AHMAD(1754) SECTION OF GENETICS 16. -
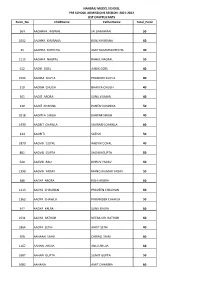
HANSRAJ MODEL SCHOOL PRE SCHOOL ADMISSIONS SESSION -2021-2022 LIST OFAPPLICANTS Form No Childname Fathername Total Point
HANSRAJ MODEL SCHOOL PRE SCHOOL ADMISSIONS SESSION -2021-2022 LIST OFAPPLICANTS Form_No ChildName FatherName Total_Point 564 AADHAYA JAISWAL JAI BHAGWAN 50 1002 AADHYA KHURANA KUNJ KHURANA 60 35 AADHYA KOTECHA AMIT KUMAR KOTECHA 40 1115 AADHYA NAGPAL RAHUL NAGPAL 50 612 AADIK GOEL AADIK GOEL 40 1554 AADIRA GUPTA PRABODH GUPTA 80 319 AADISH CHUGH BHAVYA CHUGH 40 301 AADIT ARORA SUNIL KUMAR 40 130 AADIT KHANNA PUNESH KHANNA 50 2018 AADITYA SINGH ISHWAR SINGH 40 1930 AADRIT CHAWLA SAURABH CHAWLA 60 144 AADRITI SATISH 50 1870 AADVIK GOYAL AADVIK GOYAL 40 881 AADVIK GUPTA SACHIN GUPTA 50 620 AADVIK RAO DHRUV YADAV 60 1399 AADVIK YADAV MANOJ KUMAR YADAV 50 386 AADYA ARORA RISHI ARORA 50 1213 AADYA CHAUHAN PRAVEEN CHAUHAN 60 1363 AADYA CHAWLA PARVINDER CHAWLA 50 347 AADYA KALRA SUNIL KALRA 50 1031 AADYA RATHOR NEERAJ KR. RATHOR 60 1864 AADYA SETHI ANKIT SETHI 40 508 AAHAAN SAINI CHIRAG SAINI 60 1167 AAHAN AHUJA ANUJ AHUJA 60 1687 AAHAN GUPTA SUMIT GUPTA 50 1682 AAHANA AMIT CHHABRA 60 HANSRAJ MODEL SCHOOL PRE SCHOOL ADMISSIONS SESSION -2021-2022 LIST OFAPPLICANTS Form_No ChildName FatherName Total_Point 457 AAHANA ARORA RAHUL ARORA 50 687 AAHANA DHINGRA DEEPAK DHINGRA 60 1853 AAHANA MIDHA GAURAV MIDHA 50 1700 AAHIRA VERMA NITIN VERMA 40 100 AAHNA KAUR RANJIT SINGH 40 ANUPAM KUMAR 162 AAKARSH KUMAR CHOUDHARY CHAUDHARY 40 245 AAKRISHI KOHLI RISHI RAJ KOHLI 60 177 AAMYA GANDHI AKSHIT GANDHI 50 1449 AANFATEH SINGH JOHAR NAVJOT SINGH 50 331 AANYA GUPTA LOVE KUMAR GUPTA 50 1477 AARADHYA DHARAMVIR SINGH THAGELA 60 1209 AARADHYA BOHRA DR. -

National Institutional Ranking Framework
National Institutional Ranking Framework Ministry of Human Resource Development Government of India Welcome to Data Capturing System: OVERALL Submitted Institute Data for NIRF'2020' Institute Name: ACADEMY OF SCIENTIFIC & INNOVATIVE RESEARCH [IR-O-U-0713] Sanctioned (Approved) Intake Academic Year 2018-19 2017-18 2016-17 2015-16 2014-15 2013-14 PG [1 Year Program(s)] 17 - - - - - PG [2 Year Program(s)] 136 65 - - - - Total Actual Student Strength (Program(s) Offered by Your Institution) (All programs No. of Male No. of Female Total Students Within State Outside State Outside Economically Socially No. of students No. of students No. of students No. of students of all years) Students Students (Including male (Including male Country Backward Challenged receiving full receiving full receiving full who are not & female) & female) (Including male (Including male (SC+ST+OBC tuition fee tuition fee tuition fee receiving full & female) & female) Including male reimbursement reimbursement reimbursement tuition fee & female) from the State from Institution from the Private reimbursement and Central Funds Bodies Government PG [1 Year 6 3 9 2 7 0 0 2 0 0 0 2 Program(s)] PG [2 Year 53 56 109 48 61 0 1 42 1 1 0 41 Program(s)] Placement & Higher Studies PG [1 Years Program(s)]: Placement & higher studies for previous 3 years Academic Year No. of first year No. of first year Academic Year No. of students graduating in minimum No. of students Median salary of No. of students students intake in the students admitted in stipulated time placed placed selected for Higher year the year graduates(Amount in Studies Rs.) 2016-17 20 20 2016-17 17 5 319000(Rupees Three 9 Lakhs Nineteen Thousand Only) 2017-18 16 16 2017-18 16 10 330000(Rupees Three 2 Lakhs Thirty Thousand Only) 2018-19 17 9 2018-19 9 2 325000(Rupees Three 4 Lakhs Twenty Five Thousand Only) PG [2 Years Program(s)]: Placement & higher studies for previous 3 years Academic Year No. -

Cover Legend: Gopal Kundu; a Member of the Editorial Academy of the International Journal of Oncology
INTERNATIONAL JOURNAL OF ONCOLOGY 58: MAY, 2021 Cover legend: Gopal Kundu; a member of The Editorial Academy of The International Journal of Oncology the area of research specific to disease known as ‘Coronary Artery Restenosis after angioplasty’ and published a study on this topic (Potential roles of osteopontin and avb3 integrin in the development of coronary artery restenosis after angioplasty. Proc Natl Acad Sci USA 94: 9308‑9313,1997). In addition, he filed a US patent along with his team on ‘Methods and compositions for treatment of restenosis. US Patent No. US 6,458,590 B1' in 2002 and received a Fellows Award for Research Excellence (FARE) for best research from NIH, USA in 1997. Professor Kundu subsequently returned to India in 1998 and Professor Gopal C. Kundu, was born into a very poor family acquired the position of Scientist‑D (equivalent to Assistant in a remote village of West Bengal, India. He completed his Professor) at the National Centre for Cell Science (NCCS), Pune, undergraduate (1977‑1980) and postgraduate (1980‑1983) India under the Ministry of Science and Technology, Government studies in Chemistry at the University of Calcutta, Kolkata, India of India. He was then promoted to Scientist‑E, Scientist‑F and and then obtained his Ph.D. degree in Chemistry from Bose then Scientist‑G (equivalent to Professor). He spent his valuable Institute (Sir J. C. Bose), Kolkata, India in 1989. time at NCCS, Pune, India from 1998 to 2019 and his area of Professor Kundu undertook his post‑doctoral research at research at NCCS was in the area of Tumor biology, cancer stem the Department of Heart and Hypertension (now known as the cells, tumor‑stromal interaction, angiogenesis, cancer Department of Molecular Cardiology), The Cleveland Clinic therapeutics, biomarker development and nanomedicine. -

Year Book 2018 Year Book 2018
YEAR BOOK 2018 YEAR BOOK 2018 WEST BENGAL ACADEMY OF SCIENCE AND TECHNOLOGY CSIR-Indian Institute of Chemical Biology Jadavpur YEAR BOOK Kolkata 700 032 Registered under the West Bengal Act XXVI of 1961 (S/65001 of 1990-91) 2018 PAN – AAATW0707E Published by : Prof. Satyabrata Pal, Elected Member, ISI, FRSS Formerly, Dean, Post Graduate Studies, BCKV and Honorary Visiting Professor, ISI, Kolkata Editor, West Bengal Acadepmy of Science and Technology Assisted by : Dr. Arun Bandyopadhyay, Ph.D. Chief Scientist, CSIR-IICB, Kolkata-700 032 Secretary, West Bengal Academy of Science and Technology WAST Secretariat CSIR-Indian Institute of Chemical Biology 4, Raja S. C. Mullick Road WEST BENGAL Jadavpur, Kolkata 700 032 A C Telephone: (033) 2499-5796 A W A D e-mail: [email protected] E M Website: http://www.iicb.res.in/wast/index.html S T Y SCIENCE Printed by : WEST BENGAL ACADEMY OF SCIENCE AND TECHNOLOGY Creative Data Centre Registered Office : CSIR-Indian Institute of Chemical Biology 58/32, Prince Anwar Shah Road 4, Raja S. C. Mullick Road, Jadavpur Kolkata- 700 045 Kolkata 700 032 E-mail: [email protected] 1 2 YEAR BOOK 2018 YEAR BOOK 2018 AD-HOC Committee (1986-1989) Contents 1. Professor Sushil Kumar Mukherjee : Chairman 2. Professor Syama Pada Sen Introduction 5 3. Professor Asok Ghosh Memorandum of Association 6 4. Dr. Satyesh Chandra Pakrashi Rules and Regulations 9 Approved Amendments–I 25 5. Professor Subodh Kumar Roy Approved Amendments–II 29 6. Professor Asok Kumar Barua Past Office Bearers 34 7. Professor Nityananda Saha Council : 2016-2018 37 Sectional Committees : 2016-2018 39 8. -

LIST of LIFE MEMBERS AS on 1St SEPTEMBER 2020
LIST OF LIFE MEMBERS AS ON 1st SEPTEMBER 2020 10. Dr. SHAGUFTA MOIN (1261) ALIGARH DEPT. OF BIOCHEMISTRY J. N. MEDICAL COLLEGE 1. Dr. HAMMAD AHMAD SHADAB G. G.(1454) ALIGARH MUSLIM UNIVERSITY 31 SECTOR OF GENETICS ALIGARH 202 002 DEPT. OF ZOOLOGY ALIGARH MUSLIM UNIVERSITY 11. SHAIK NISAR ALI (3769) ALIGARH 202 002 DEPT. OF BIOCHEMISTRY FACULTY OF LIFE SCIENCE 2. Dr. INDU SAXENA (1838) ALIGARH MUSLIM UNIVERSITY HIG 30, ADA COLONY ALIGARH 202 002 AVANTEKA PHASE I [email protected] RAMGHAT ROAD, ALIGARH 202 001 12. DR. MAHAMMAD REHAN AJMAL KHAN (4157) 3. Dr. (MRS) KHUSHTAR ANWAR SALMAN(3332) 4/570, Z-5, NOOR MANZIL COMPOUND DEPT. OF BIOCHEMISTRY DIDHPUR, CIVIL LINES JAWAHARLAL NEHRU MEDICAL COLLEGE ALIGARH UP 202 002 ALIGARH MUSLIM UNIVERSITY [email protected] ALIGARH 202 002 [email protected] 13. DR. HINA YOUNUS (4281) INTERDISCIPLINARY BIOTECHNOLOGY UNIT 4. Dr. MOHAMMAD TABISH (2226) ALIGARH MUSLIM UNIVERSITY DEPT. OF BIOCHEMISTRY ALIGARH U.P. 202 002 FACULTY OF LIFE SCIENCES [email protected] ALIGARH MUSLIM UNIVERSITY ALIGARH 202 002 [email protected] ALLAHABAD 5. Dr. MOHAMMAD AFZAL (1101) 14. Dr. B. SHARMA (872) DEPT. OF ZOOLOGY DEPARTMENT OF BIOCHEMISTRY ALIGARH MUSLIM UNIVERSITY UNIVERSITY OF ALLAHABAD ALIGARH 202 002 ALLAHABAD 211002 INDIA [email protected] 6. Dr. RIAZ AHMAD(1754) [email protected] SECTION OF GENETICS 9415715639 DEPT. OF ZOOLOGY ALIGARH MUSLIM UNIVERSITY 15. Dr. ABHAY KUMAR PANDEY (2054) ALIGARH 202 002 DEPT OF BIOCHEMISTRY UNIVERSITY OF ALLAHABAD 7. Dr. ROSHAN ALAM (1958) ALLAHABAD 211002 C/O MOHD HABIB EDUCATIONAL HOUSE AMU SHAM SHAD MARKET 16. -

Awardees of National Bioscience Award for Career Development
AWARDEES OF NATIONAL BIOSCIENCE AWARD FOR CAREER DEVELOPMENT Awardees for the year 2016 1. Dr. Mukesh Jain, Associate Professor, School of Computational and Integrative Sciences, Jawaharlal Nehru University, New Delhi-110067 2. Dr. Samir K. Maji, Associate Professor, Indian Institute of Technology, Powai, Mumbai- 400076 3. Dr. Anindita Ukil, Assistant Professor, Calcutta University, Kolkata 4. Dr. Arnab Mukhopadhyay, Staff Scientist V, National Institute of Immunology, Aruna Asaf Ali Marg, New Delhi- 110067 5. Dr. Rohit Srivastava, Professor, Indian Institute of Technology, Bombay, Mumbai- 400076 6. Dr. Pinaki Talukdar, Associate Professor, Indian Institute of Science Education and Research, Dr. Homi Bhabha Road, Pashan, Pune- 7. Dr. Rajnish Kumar Chaturvedi, Senior Scientist, CSIR- Indian Institute of Toxicology Research, Lucknow-226001 8. Dr. Jackson James, Scientist E-II, Neuro Stem Cell Biology Lab, Neurobiology Division, Rajiv Gandhi Centre for Biotechnology, Thiruvananthapuram, Kerala- 695014 Awardees for the year 2015 1. Dr. Sanjeev Das, Staff Scientist-V, National Institute of Immunology, New Delhi 2. Dr. Ganesh Nagaraju, Assistant Professor, Department of Biotechnology, Indian Institute of Science, Bangalore- 5600012. 3. Dr. Suvendra Nath Bhattacharya, Principal Scientist, CSIR- Indian Institute of Chemical Biology, Kolkata- 700032 4. Dr. Thulasiram H V, Principal Scientist, CSIR-National Chemical Laboratory, Pune- 411008. 5. Dr. Pawan Gupta, Principal Scientist, Institute of microbial Technology, Chandigarh- 160036. 6. Dr. Souvik Maiti, Principal Scientist, CSIR-Institute of Genomics and Integrative Biology, Delhi- 110025. 7. Dr. Pravindra Kumar, Associate Professor, Department of Biotechnology, IIT, Roorkee- 247667. 8. Dr. Anurag Agrawal, Principal Scientist, CSIR-Institute of Genomics and Integrative Biology, Delhi- 110025 9. Dr. Gridhar Kumar Pandey, Professor, Department of Plant Molecular Biology, University of Delhi South Campus, New Delhi- 110067 10. -

Insights from a Pan India Sero-Epidemiological Survey (Phenome-India Cohort) for SARS-Cov-2
medRxiv preprint doi: https://doi.org/10.1101/2021.01.12.21249713; this version posted February 28, 2021. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted medRxiv a license to display the preprint in perpetuity. It is made available under a CC-BY-NC-ND 4.0 International license . Title Page Insights from a Pan India Sero-Epidemiological survey (Phenome-India Cohort) for SARS-CoV-2 Salwa Naushin, MSc1,32#; Viren Sardana, MBBS, M.Tech1,32#; Rajat Ujjainiya, MSc1,32; Nitin Bhatheja, BTech1; Rintu Kutum, MSc1,32; Akash Kumar Bhaskar, MSc1,32; Shalini Pradhan, MSc1; Satyartha Prakash, MTech1; Raju Khan, PhD2,32; Birendra Singh Rawat, PhD3,32; Karthik Bharadwaj Tallapaka, MD4; Mahesh Anumalla, MSc4; Giriraj Ratan Chandak, PhD4,32 ; Amit Lahiri, PhD5,32; Susanta Kar, PhD5,32; Shrikant Ramesh Mulay,PhD5,32; Madhav Nilakanth Mugale, PhD5,32; Mrigank Srivastava, PhD5,32; Shaziya Khan, MSc5,32; Anjali Srivastava, MSc5,32; Bhawna Tomar, MSc5,32; Murugan Veerapandian, PhD6,32; Ganesh Venkatachalam, PhD6,32; Selvamani Raja Vijayakumar, MBBS6; Ajay Agarwal, PhD7,32; Dinesh Gupta, MD7; Prakash M Halami, PhD8,32; Muthukumar Serva Peddha, PhD8,32; Gopinath M Sundaram, PhD8,32; Ravindra P Veeranna, PhD8,32; Anirban Pal, PhD9,32; Vinay Kumar Agarwal, MD9; Anil Ku Maurya, PhD9; Ran Vijay Kumar Singh, PhD10,32; Ashok Kumar Raman, MBBS10; Suresh Kumar Anandasadagopan, PhD11,32; Parimala Karuppanan, MBBS11; Subramanian Venkatesan, PhD11,32; Harish Kumar Sardana, PhD12; Anamika Kothari, MBBS12; -

Carriage & Wagon Workshop/ Eastern Railway / Liluah
CARRIAGE & WAGON WORKSHOP/ EASTERN RAILWAY / LILUAH ELECTRORAL ROLL FOR SECRET BALLOT ELECTION - 2020 S/NO. EMP. NO. TICKET NO. EMPLOYEE NAME DESIGNATION BILL UNIT SHOP/OFFICE REMARKS BOOTH NO. 1 03629800018 10300077 SAIKAT MONDAL TECH.(FITTER)-III 0206101 E-SHOP Booth No. 01 2 03629800071 10200095 BISHNU NATH TECH.(WELDER)-III 0206101 C-SHOP Booth No. 01 3 03629800239 10204430 KISHORE SAHA TECH.(FITTER)-III 0206101 C-SHOP Booth No. 01 4 03629800241 10204441 SMARAJIT MAZUMDER TECH.(FITTER)-III 0206101 C-SHOP Booth No. 01 5 03629800242 10204453 SURAJIT BISWAS TECH.(FITTER)-III 0206101 C-SHOP Booth No. 01 6 03629800243 10204465 RAJIB BISWAS TECH.(FITTER)-III 0206101 C-SHOP Booth No. 01 7 03629800244 10300089 SAMSUR BISWAS TECH.(FITTER)-III 0206101 E-SHOP Booth No. 01 8 03629800245 10103976 DHANANJOY MISTRY TECH.(FITTER)-III 0206101 A-SHOP Booth No. 01 9 03629800246 10104045 SAJAL DAS TECH.(FITTER)-III 0206101 A-SHOP Booth No. 01 10 03629800249 10204477 RATAN HALDER TECH.(FITTER)-III 0206101 C-SHOP Booth No. 01 11 03629800250 10300211 BIJAY GOUR TECH.(FITTER)-III 0206101 E-SHOP Booth No. 01 12 03629800251 10104197 JALDHAR KUMAR TECH.(FITTER)-III 0206101 A-SHOP Booth No. 01 13 03629800285 10300193 ADITYA KUMAR PASWAN TECH(MACHINIST)-III 0206101 E-SHOP Booth No. 01 14 50200039123 10104112 ASIT KUMAR BISWAS TECH.(FITTER)-I 0206101 A-SHOP Booth No. 01 15 50200063484 12300846 DIPAK PRASAD TECH.(WELDER)-I 0206101 . Booth No. 01 16 50202265527 10201750 KRISHNA YADAV TECH.(FITTER)-I 0206101 C-SHOP Booth No. 01 17 50202265576 10402834 TARAK NATH DAS TECH.(FITTER)-I 0206101 F-SHOP Booth No. -

Book Download
SOCIETY OF BIOLOGICAL CHEMISTS (INDIA) (1930 – 2011) 1 TABLE OF CONTENTS 1. Goals and activities of SBC(I) 2. Rules and Bye-laws of SBC(I) 3. Past Presidents, Secretaries, Treasurers (with tenure) 4. “Reminiscences on the development of the Society of Biological Chemists (India): a personal perspective” by Prof. N. Appaji Rao 5. “Growth of Biochemistry in India” by Prof. G. Padmanaban 6. Current office bearers 7. Current Executive Committee Members 8. Office staff 9. Past meeting venues of SBC(I) 10. SBC(I) awards, criteria and procedure for applying 11. SBC(I) awardees 12. Current list of life members with address 13. Acknowledgments 2 GOALS AND ACTIVITIES OF SBC(I) To meet a long felt need of scientists working in the discipline of biological chemistry " The Society Of Biological Chemists (India)" was founded in 1930, with its Head Quarters at Indian Institute of Science, Bangalore. It was registered under the Societies Act in the then princely state of Mysore and the memorandum of registration was signed by the late Profs. V. Subramanian, V. N. Patwardhan and C. V. Natarajan, who were leading personalities in the scientific firmament during that period. The Society played a crucial role during the Second World War by advising the Government on the utilization of indigenous biomaterials as food substitutes, drugs and tonics, on the industrial and agricultural waste utilization and on management of water resources. The other areas of vital interest to the Society in the early years were nutrition, proteins, enzymes, applied microbiology, preventive medicines and the development of high quality proteins from indigenous plant sources.