Supporting Information for Proteomics DOI 10.1002/Pmic.200500652
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Associated Palmoplantar Keratoderma
DR ABIGAIL ZIEMAN (Orcid ID : 0000-0001-8236-207X) Article type : Review Article Pathophysiology of pachyonychia congenita-associated palmoplantar keratoderma: New insight into skin epithelial homeostasis and avenues for treatment Authors: A. G. Zieman1 and P. A. Coulombe1,2 # Affiliations: 1Department of Cell and Developmental Biology, University of Michigan Medical School, Ann Arbor, MI 48109, USA; 2Department of Dermatology, University of Michigan Medical School, Ann Arbor, MI 48109, USA #Corresponding author: Pierre A. Coulombe, PhD, 3071 Biomedical Sciences Research Building, 109 Zina Pitcher Place, Ann Arbor, MI 48109, USA. Tel: 734-615-7509. Email: [email protected]. Funding Sources: These studies were supported by grant AR044232 issued to P.A.C. from the National Institute of Arthritis, Musculoskeletal and Skin Disease (NIAMS). A.G.Z. received support from grant T32 CA009110 from the National Cancer Institute. Author Manuscript This is the author manuscript accepted for publication and has undergone full peer review but has not been through the copyediting, typesetting, pagination and proofreading process, which may lead to differences between this version and the Version of Record. Please cite this article as doi: 10.1111/BJD.18033 This article is protected by copyright. All rights reserved Conflict of interest disclosures: None declared. Bulleted statements: What’s already known about this topic? Pachyonychia congenita is a rare genodermatosis caused by mutations in KRT6A, KRT6B, KRT6C, KRT16, KRT17, which are normally expressed in skin appendages and induced following injury. Individuals with PC present with multiple clinical symptoms that usually include thickened and dystrophic nails, palmoplantar keratoderma (PPK), glandular cysts, and oral leukokeratosis. -

Mass Spectrometry-Based Proteomics Techniques and Their Application in Ovarian Cancer Research Agata Swiatly, Szymon Plewa, Jan Matysiak and Zenon J
Swiatly et al. Journal of Ovarian Research (2018) 11:88 https://doi.org/10.1186/s13048-018-0460-6 REVIEW Open Access Mass spectrometry-based proteomics techniques and their application in ovarian cancer research Agata Swiatly, Szymon Plewa, Jan Matysiak and Zenon J. Kokot* Abstract Ovarian cancer has emerged as one of the leading cause of gynecological malignancies. So far, the measurement of CA125 and HE4 concentrations in blood and transvaginal ultrasound examination are essential ovarian cancer diagnostic methods. However, their sensitivity and specificity are still not sufficient to detect disease at the early stage. Moreover, applied treatment may appear to be ineffective due to drug-resistance. Because of a high mortality rate of ovarian cancer, there is a pressing need to develop innovative strategies leading to a full understanding of complicated molecular pathways related to cancerogenesis. Recent studies have shown the great potential of clinical proteomics in the characterization of many diseases, including ovarian cancer. Therefore, in this review, we summarized achievements of proteomics in ovarian cancer management. Since the development of mass spectrometry has caused a breakthrough in systems biology, we decided to focus on studies based on this technique. According to PubMed engine, in the years 2008–2010 the number of studies concerning OC proteomics was increasing, and since 2010 it has reached a plateau. Proteomics as a rapidly evolving branch of science may be essential in novel biomarkers discovery, therapy decisions, progression predication, monitoring of drug response or resistance. Despite the fact that proteomics has many to offer, we also discussed some limitations occur in ovarian cancer studies. -

Keratin 9 Point Mutation in the Pedigree of Epidermolytic Hereditary Palmoplantar Keratoderma Perturbs Keratin Intermediate Filament Network Formation
FEBS 17004 FEBS Letters 386 (1996) 149-155 Keratin 9 point mutation in the pedigree of epidermolytic hereditary palmoplantar keratoderma perturbs keratin intermediate filament network formation Setsu Kobayashi, Toshihiro Tanaka*, Norihisa Matsuyoshi, Sadao Imamura Department of Dermatology, Graduate School of Medicine, Kyoto University, Kyoto, 606 Japan Received 12 January 1996; revised version received 4 April 1996 Abstract Keratins form an intracellular keratin filament net- point mutations in the K9 gene in EHPPK [4-8] but none work in keratinocytes. Point mutations in the epidermal keratins showed a function assay with these mutations. Here, we pro- could lead to the disruption of keratin filament formation, vide the first demonstration that the point mutation found in developing skin diseases such as epidermolytic hereditary a pedigree of EHPPK has a dominant-negative effect on the palmoplantar keratoderma (EHPPK). We found a G to A assembly of keratin intermediate filaments in the cells. transition in keratin 9 (K9) cDNA, resulting in the substitution of glutamine for arginine at 162, in all patients of a pedigree of 2. Materials and methods EHPPK. Transfection into MDCK cells and DJM-1 cells revealed that the plasmid CMX vector containing normal keratin 2.1. PCR and DNA sequence 9 cDNA showed normal keratin network formation, whereas the Genomic DNA was extracted and purified from blood or biopsy vector with a G to A point mutated keratin 9 cDNA showed specimens from the patients. The primers were designed at nucleotide disrupted keratin filaments with droplet formation in the cells. 263 282 and 664-683 based on the K9 cDNA sequence [9]. -

Comparative Genomics Analyses of Alpha-Keratins Reveal Insights Into
Sun et al. Frontiers in Zoology (2017) 14:41 DOI 10.1186/s12983-017-0225-x RESEARCH Open Access Comparative genomics analyses of alpha- keratins reveal insights into evolutionary adaptation of marine mammals Xiaohui Sun, Zepeng Zhang, Yingying Sun, Jing Li, Shixia Xu* and Guang Yang* Abstract Background: Diversity of hair in marine mammals was suggested as an evolutionary innovation to adapt aquatic environment, yet its genetic basis remained poorly explored. We scanned α-keratin genes, one major structural components of hair, in 16 genomes of mammalian species, including seven cetaceans, two pinnipeds, polar bear, manatee and five terrestrial species. Results: Extensive gene loss and high pseudogenization rate of α-keratin genes were identified in cetaceans when compared to terrestrial artiodactylans (average number of α-keratins 37.29 vs. 58.33; pseudogenization rate 29.89% vs. 8.00%), especially of hair follicle-specific keratin genes (average pseudogenization rate in cetaceans of 43.88% relative to 3.80% artiodactylian average). Compared to toothed whale, the much more number of intact functional α-keratin genes was examined in the baleen whale that had specific keratinized baleen. In contrast, the number of keratin genes in pinnipeds, polar bear and manatee were comparable to those of their respective terrestrial relatives. Additionally, four keratin genes (K39, K9, K42, and K74) were found to be pseudogenes or lost uniquely in cetaceans and manatees. Conclusions: Species-specific evolution of α-keratin gene family identified in the marine mammals might be responsible for their different hair characteristics. Increased gene loss and pseudogenization rate identified in cetacean lineages was likely to contribute to hair-less phenotype to adaptation for complete aquatic environment. -
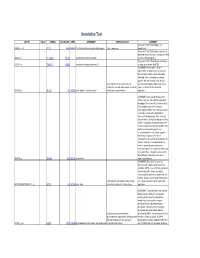
Supplementary Table 5. Functional Annotation of the Largest Gene Cluster(221 Element)
Annotation Tool AFFYID VALUE SYMBOL LOCUSLINK OMIM GENENAME GENEONTOLOGY SUMMARY [Proteome FUNCTION:] Expressed 203054_s_at TCTA 6988 600690 T-cell leukemia translocation altered gene tumor suppressor ubiquitously [Proteome FUNCTION:] May be involved in protein-protein interactions; contains five WD 44563_at FLJ10385 55135 hypothetical protein FLJ10385 domains (WD-40 repeats) [Proteome FUNCTION:] Weakly similarity to 212261_at TNRC15 26058 trinucleotide repeat containing 15 a region of rat nestin (Rn.9701) [SUMMARY:] Actin alpha 1 which is expressed in skeletal muscle is one of six different actin isoforms which have been identified. Actins are highly conserved proteins that are involved in cell motility, actin filament; motor activity; muscle structure and integrity. Alpha actins are a contraction; muscle development; structural major constituent of the contractile 203872_at ACTA1 58 102610 actin, alpha 1, skeletal muscle constituent of cytoskeleton apparatus. [SUMMARY:] Annexin VIII belong to the family of Ca (2+) dependent phospholipid binding proteins (annexins), and has a high 56% identity to annexin V (vascular anticoagulant-alpha). It was initially isolated as 2.2 kb vascular anticoagulant-beta transcript from human placenta, a Ca (2+) dependent phospholipid binding protein that inhibits coagulation and phospholipase A2 activity. However, the fact that annexin VIII is neither an extracellular protein nor associated with the cell surface suggests that it may not play a role in blood coagulation in vivo and its physiological role remains unknown. It is expressed at low levels in human placenta and shows restricted expression in lung endothelia, skin, liver, and kidney. The gene is also found to be selectively overexpressed in acute 203074_at ANXA8 244 602396 annexin A8 myelocytic leukemia. -

Proteomic Approaches Identify Members of Cofilin Pathway Involved in Oral Tumorigenesis
Proteomic Approaches Identify Members of Cofilin Pathway Involved in Oral Tumorigenesis Giovana M. Polachini1, Lays M. Sobral2, Ana M. C. Mercante3, Adriana F. Paes-Leme4, Fla´via C. A. Xavier5, Tiago Henrique1, Douglas M. Guimara˜es6, Alessandra Vidotto1, Erica E. Fukuyama7, Jose´ F. Go´ is-Filho7, Patricia M. Cury8, Ota´vio A. Curioni9, Pedro Michaluart Jr10, Adriana M. A. Silva11, Victor Wu¨ nsch-Filho12, Fabio D. Nunes6, Andre´ia M. Leopoldino2, Eloiza H. Tajara1,13* 1 Departamento de Biologia Molecular; Faculdade de Medicina (FAMERP), Sa˜oJose´ do Rio Preto, SP, Brazil, 2 Departamento de Ana´lises Clı´nicas, Toxicolo´gicas e Bromatolo´gicas, Faculdade de Cieˆncias Farmaceˆuticas da Universidade de Sa˜o Paulo, Ribeira˜o Preto, SP, Brazil, 3 Laborato´rio de Patologia, Hospital Helio´polis, Sa˜o Paulo, SP, Brazil, 4 Laborato´rio Nacional de Biocieˆncias (LNBio), Centro Nacional de Pesquisa em Energia e Materiais, Campinas, SP, Brazil, 5 Departamento de Propedeˆutica e Clı´nica Integrada, Faculdade de Odontologia da Universidade Federal da Bahia, Salvador,BA, Brazil, 6 Departamento de Estomatologia, Faculdade de Odontologia da Universidade de Sa˜o Paulo, Sa˜o Paulo, SP, Brazil, 7 Servic¸o de Cirurgia de Cabec¸a e Pescoc¸o, Instituto do Caˆncer Arnaldo Vieira de Carvalho, Sa˜o Paulo, SP, Brazil, 8 Departamento de Patologia e Medicina Legal, Faculdade de Medicina (FAMERP), Sa˜oJose´ do Rio Preto, SP, Brazil, 9 Departamento de Cirurgia de Cabec¸a e Pescoc¸o e Otorrinolaringologia, Hospital Helio´polis, Sa˜o Paulo, SP, Brazil, 10 Divisa˜o -

The Correlation of Keratin Expression with In-Vitro Epithelial Cell Line Differentiation
The correlation of keratin expression with in-vitro epithelial cell line differentiation Deeqo Aden Thesis submitted to the University of London for Degree of Master of Philosophy (MPhil) Supervisors: Professor Ian. C. Mackenzie Professor Farida Fortune Centre for Clinical and Diagnostic Oral Science Barts and The London School of Medicine and Dentistry Queen Mary, University of London 2009 Contents Content pages ……………………………………………………………………......2 Abstract………………………………………………………………………….........6 Acknowledgements and Declaration……………………………………………...…7 List of Figures…………………………………………………………………………8 List of Tables………………………………………………………………………...12 Abbreviations….………………………………………………………………..…...14 Chapter 1: Literature review 16 1.1 Structure and function of the Oral Mucosa……………..…………….…..............17 1.2 Maintenance of the oral cavity...……………………………………….................20 1.2.1 Environmental Factors which damage the Oral Mucosa………. ….…………..21 1.3 Structure and function of the Oral Mucosa ………………...….……….………...21 1.3.1 Skin Barrier Formation………………………………………………….……...22 1.4 Comparison of Oral Mucosa and Skin…………………………………….……...24 1.5 Developmental and Experimental Models used in Oral mucosa and Skin...……..28 1.6 Keratinocytes…………………………………………………….….....................29 1.6.1 Desmosomes…………………………………………….…...............................29 1.6.2 Hemidesmosomes……………………………………….…...............................30 1.6.3 Tight Junctions………………………….……………….…...............................32 1.6.4 Gap Junctions………………………….……………….….................................32 -

Proteome Profiling of Exosomes Purified from a Small Amount Of
proteomes Article Proteome Profiling of Exosomes Purified from a Small Amount of Human Serum: The Problem of Co-Purified Serum Components Mateusz Smolarz 1, Monika Pietrowska 1, Natalia Matysiak 2, Łukasz Miela ´nczyk 2 and Piotr Widłak 1,* 1 Maria Skłodowska-Curie Institute—Oncology Center, Gliwice Branch, 44-101 Gliwice, Poland; [email protected] (M.S.); [email protected] (M.P.) 2 Department of Histology and Cell Pathology, School of Medicine with Division of Dentistry in Zabrze, Medical University of Silesia, 41-800 Zabrze, Poland; [email protected] (N.M.); [email protected] (Ł.M.) * Correspondence: [email protected]; Tel.: +48-32-2789672 Received: 26 March 2019; Accepted: 26 April 2019; Published: 28 April 2019 Abstract: Untargeted proteomics analysis of extracellular vesicles (EVs) isolated from human serum or plasma remains a technical challenge due to the contamination of these vesicles with lipoproteins and other abundant serum components. Here we aimed to test a simple method of EV isolation from a small amount of human serum (<1 mL) using the size-exclusion chromatography (SEC) standalone for the discovery of vesicle-specific proteins by the untargeted LC–MS/MS shotgun approach. We selected the SEC fraction containing vesicles with the size of about 100 nm and enriched with exosome markers CD63 and CD81 (but not CD9 and TSG101) and analyzed it in a parallel to the subsequent SEC fraction enriched in the lipoprotein vesicles. In general, there were 267 proteins identified by LC–MS/MS in exosome-containing fraction (after exclusion of immunoglobulins), yet 94 of them might be considered as serum proteins. -

Identification of Trichoplein, a Novel Keratin Filament- Binding Protein
Research Article 1081 Identification of trichoplein, a novel keratin filament- binding protein Miwako Nishizawa1,*, Ichiro Izawa1,*, Akihito Inoko1,*, Yuko Hayashi1, Koh-ichi Nagata1, Tomoya Yokoyama1,2, Jiro Usukura3 and Masaki Inagaki1,‡ 1Division of Biochemistry, Aichi Cancer Center Research Institute, 1-1 Kanokoden, Chikusa-ku, Nagoya 464-8681, Japan 2Department of Dermatology, Mie University Faculty of Medicine, 2-174 Edobashi, Tsu 514-8507, Japan 3Department of Anatomy and Cell Biology, Nagoya University School of Medicine, 65 Tsurumai, Showa-ku, Nagoya 466-8550, Japan *These authors contributed equally to this work ‡Author for correspondence (e-mail: [email protected]) Accepted 29 November 2004 Journal of Cell Science 118, 1081-1090 Published by The Company of Biologists 2005 doi:10.1242/jcs.01667 Summary Keratins 8 and 18 (K8/18) are major components of the antibody in a complex with K8/18 and immunostaining intermediate filaments (IFs) of simple epithelia. We report revealed that trichoplein colocalized with K8/18 filaments here the identification of a novel protein termed in HeLa cells. In polarized Caco-2 cells, trichoplein trichoplein. This protein shows a low degree of sequence colocalized not only with K8/18 filaments in the apical similarity to trichohyalin, plectin and myosin heavy chain, region but also with desmoplakin, a constituent of and is a K8/18-binding protein. Among interactions desmosomes. In the absorptive cells of the small intestine, between trichoplein and various IF proteins that we trichoplein colocalized with K8/18 filaments at the apical tested using two-hybrid methods, trichoplein interacted cortical region, and was also concentrated at desmosomes. -

Vimentin, Carcinoembryonic Antigen and Keratin in the Diagnosis of Mesothelioma, Adenocarcinoma and Reactive Pleural Lesions
Eur Respir J 1990, 3, 997-1001 Vimentin, carcinoembryonic antigen and keratin in the diagnosis of mesothelioma, adenocarcinoma and reactive pleural lesions N. AI-Saffar, P .S. Hasleton Vimentin, carcinoembryonic antigen and keratin in the diagnosis of Dept of Pathology, Regional CardiO!horacic Centre, mesothelioma, adenocarcinoma and reactive pleural lesions. N. Al-Saffar, P .S. Wythenshawe Hospital, Manchester, UK. Hasleton. ABSTRACT: An immunohistochemical study of reactive pleural lesions, Correspondence: P.S. Hasleton, Dept of Pathology, Wythensbawe Hospital, Southmoor Road, adenocarcinomas and mesothellomas using carclnoembyronic antigen Wythenshawe, Manchester M23 9LT, UK. (CEA), cytokeratln and vlmentln was carried out. All the specimens were obtained at surgery except for 11 mesotheliomas found at necropsy. Keywords: Adenocarcinoma (lung); carcinoembryonic Vlmentln was positive In 23 or 27 mesotheliomas and negative In all the antigen (CEA); keratin; mesothelioma (pleural); adenocarcinomas and 4 of 17 reactive mesothelial lesions. Conversely, vimentin. CEA was positive In all the adenocarcinomas but negative 1n all mesothe liomas. Immunoreactivity for vlmentln was seen In only 3 of 11 post Received: February 1990; accepted after revision May mortem mesotheliomas. Vlmentln Is a useful adjunct to the tissue diagnosis 2, 1990. of mesothelioma especially when CEA Is negative and cytokeratln positive. Its use appears largely confirmed to well fixed surgically derived tissues. Eur Respir J., 1990, 3, 997- 1001. The diagnosis of malignant pleural mesothelioma is 38 mesothelioma cases were necropsy specimens often difficult. It may be confused histologically with a coming mainly from the Medical Boarding Panel (M.B.P.) reactive pleurisy or adenocarcinoma. Separation from (Respiratory Diseases). Seven were epithelial, three benign lesions is obviously important. -

Clinical Chemistry
June 1 2007, Volume 53, Issue 6,pp.999- 1180 Editorials Andrew McCaddon and Peter R. Hudson Methylation and Phosphorylation: A Tangled Relationship? Clin Chem 2007 53: 999-1000. Bob Palais Quantitative Heteroduplex Analysis Clin Chem 2007 53: 1001-1003. Eleftherios P. Diamandis Oncopeptidomics: A Useful Approach for Cancer Diagnosis? Clin Chem 2007 53: 1004-1006. Roger D. Klein The Pain Protective Haplotype: Introducing the Modern Genetic Test Clin Chem 2007 53: 1007-1009. Molecular Diagnostics and Genetics Jörn Lötsch, Inna Belfer, Anja Kirchhof, Bikash K. Mishra, Mitchell B. Max, Alexandra Doehring, Michael Costigan, Clifford J. Woolf, Gerd Geisslinger, and Irmgard Tegeder Reliable Screening for a Pain-Protective Haplotype in the GTP Cyclohydrolase 1 Gene (GCH1) Through the Use of 3 or Fewer Single Nucleotide Polymorphisms Clin Chem 2007 53: 1010-1015. Published online March 15, 2007; 10.1373/clinchem.2006.082883 Kerstin L. Edlefsen, Jonathan F. Tait, Mark H. Wener, and Michael Astion Utilization and Diagnostic Yield of Neurogenetic Testing at a Tertiary Care Facility Clin Chem 2007 53: 1016-1022. Published online April 19, 2007; 10.1373/clinchem.2006.083360 Sara Bremer, Helge Rootwelt, and Stein Bergan Real-Time PCR Determination of IMPDH1 and IMPDH2 Expression in Blood Cells Clin Chem 2007 53: 1023-1029. Published online April 26, 2007; 10.1373/clinchem.2006.081968 Elizabeth Herness Peters, Sandra Rojas-Caro, Mitchell G. Brigell, Robert J. Zahorchak, Shelley Ann des Etages, Patricia L. Ruppel, Charles R. Knight, Bradley Austermiller, Myrna C. Graham, Steve Wowk, Sean Banks, Lakshmi V. Madabusi, Patrick Turk, Donna Wilder, Carole Kempfer, Terry W. Osborn, and James C. -

Multifaceted Role of Keratins in Epithelial Cell Differentiation and Transformation
J Biosci (2019) 44:33 Ó Indian Academy of Sciences DOI: 10.1007/s12038-019-9864-8 (0123456789().,-volV)(0123456789().,-volV) Review Multifaceted role of keratins in epithelial cell differentiation and transformation 1,2 1 1,2 1,2 CRISMITA DMELLO ,SAUMYA SSRIVASTAVA ,RICHA TIWARI ,PRATIK RCHAUDHARI , 1,2 1,2 SHARADA SAWANT and MILIND MVAIDYA * 1Vaidya Laboratory, Advanced Centre for Treatment, Research and Education in Cancer (ACTREC), Tata Memorial Centre (TMC), Kharghar, Navi Mumbai 410210, India 2Homi Bhabha National Institute, Training School Complex, Anushakti Nagar, Mumbai 400085, India *Corresponding author (Email, [email protected]) MS received 18 September 2018; accepted 19 December 2018; published online 8 April 2019 Keratins, the epithelial-predominant members of the intermediate filament superfamily, are expressed in a pairwise, tissue- specific and differentiation-dependent manner. There are 28 type I and 26 type II keratins, which share a common structure comprising a central coiled coil a-helical rod domain flanked by two nonhelical head and tail domains. These domains harbor sites for major posttranslational modifications like phosphorylation and glycosylation, which govern keratin function and dynamics. Apart from providing structural support, keratins regulate various signaling machinery involved in cell growth, motility, apoptosis etc. However, tissue-specific functions of keratins in relation to cell proliferation and differ- entiation are still emerging. Altered keratin expression pattern during and after malignant transformation is reported to modulate different signaling pathways involved in tumor progression in a context-dependent fashion. The current review focuses on the literature related to the role of keratins in the regulation of cell proliferation, differentiation and transfor- mation in different types of epithelia.