Male-Biased Autosomal Effect of 16P13.11 Copy Number Variation in Neurodevelopmental Disorders
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Protein Interaction Network of Alternatively Spliced Isoforms from Brain Links Genetic Risk Factors for Autism
ARTICLE Received 24 Aug 2013 | Accepted 14 Mar 2014 | Published 11 Apr 2014 DOI: 10.1038/ncomms4650 OPEN Protein interaction network of alternatively spliced isoforms from brain links genetic risk factors for autism Roser Corominas1,*, Xinping Yang2,3,*, Guan Ning Lin1,*, Shuli Kang1,*, Yun Shen2,3, Lila Ghamsari2,3,w, Martin Broly2,3, Maria Rodriguez2,3, Stanley Tam2,3, Shelly A. Trigg2,3,w, Changyu Fan2,3, Song Yi2,3, Murat Tasan4, Irma Lemmens5, Xingyan Kuang6, Nan Zhao6, Dheeraj Malhotra7, Jacob J. Michaelson7,w, Vladimir Vacic8, Michael A. Calderwood2,3, Frederick P. Roth2,3,4, Jan Tavernier5, Steve Horvath9, Kourosh Salehi-Ashtiani2,3,w, Dmitry Korkin6, Jonathan Sebat7, David E. Hill2,3, Tong Hao2,3, Marc Vidal2,3 & Lilia M. Iakoucheva1 Increased risk for autism spectrum disorders (ASD) is attributed to hundreds of genetic loci. The convergence of ASD variants have been investigated using various approaches, including protein interactions extracted from the published literature. However, these datasets are frequently incomplete, carry biases and are limited to interactions of a single splicing isoform, which may not be expressed in the disease-relevant tissue. Here we introduce a new interactome mapping approach by experimentally identifying interactions between brain-expressed alternatively spliced variants of ASD risk factors. The Autism Spliceform Interaction Network reveals that almost half of the detected interactions and about 30% of the newly identified interacting partners represent contribution from splicing variants, emphasizing the importance of isoform networks. Isoform interactions greatly contribute to establishing direct physical connections between proteins from the de novo autism CNVs. Our findings demonstrate the critical role of spliceform networks for translating genetic knowledge into a better understanding of human diseases. -

The NDE1 Genomic Locus Affects Treatment of Psychiatric Illness Through Gene Expression Changes Related to Microrna-484
bioRxiv preprint doi: https://doi.org/10.1101/087007; this version posted November 10, 2016. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. It is made available under aCC-BY-NC-ND 4.0 International license. The NDE1 Genomic Locus Affects Treatment of Psychiatric Illness through Gene Expression Changes Related to MicroRNA-484 Nicholas J. Bradshaw1, Maiju Pankakoski2, Liisa Ukkola-Vuoti2,3, Amanda B. Zheutlin4, Alfredo Ortega-Alonso2,3, Minna Torniainen-Holm2,3, Vishal Sinha2,3, Sebastian Therman2, Tiina Paunio5,6, Jaana Suvisaari2, Jouko Lönnqvist2,5, Tyrone D. Cannon4, Jari Haukka2,7, William Hennah2,3 * 1, Department of Neuropathology, Heinrich Heine University, Düsseldorf, Germany 2, Department of Health, Mental Health Unit, National Institute for Health and Welfare, Helsinki, Finland 3, Institute for Molecular Medicine Finland FIMM, University of Helsinki, Finland 4, Department of Psychology, Yale University, USA 5, Department of Psychiatry, University of Helsinki and Helsinki University Hospital, Finland 6, Department of Health, Genomics and Biomarkers Unit, National Institute for Health and Welfare, Helsinki, Finland 7, Department of Public Health, Clinicum, University of Helsinki, Finland * Corresponding Author: William Hennah PhD, Institute for Molecular Medicine Finland FIMM, P.O. Box 20, FI-00014 University of Helsinki, Finland Email: [email protected] Tel: +358 (0)503183423 bioRxiv preprint doi: https://doi.org/10.1101/087007; this version posted November 10, 2016. The copyright holder for this preprint (which was not certified by peer review) is the author/funder, who has granted bioRxiv a license to display the preprint in perpetuity. -

Association of Gene Ontology Categories with Decay Rate for Hepg2 Experiments These Tables Show Details for All Gene Ontology Categories
Supplementary Table 1: Association of Gene Ontology Categories with Decay Rate for HepG2 Experiments These tables show details for all Gene Ontology categories. Inferences for manual classification scheme shown at the bottom. Those categories used in Figure 1A are highlighted in bold. Standard Deviations are shown in parentheses. P-values less than 1E-20 are indicated with a "0". Rate r (hour^-1) Half-life < 2hr. Decay % GO Number Category Name Probe Sets Group Non-Group Distribution p-value In-Group Non-Group Representation p-value GO:0006350 transcription 1523 0.221 (0.009) 0.127 (0.002) FASTER 0 13.1 (0.4) 4.5 (0.1) OVER 0 GO:0006351 transcription, DNA-dependent 1498 0.220 (0.009) 0.127 (0.002) FASTER 0 13.0 (0.4) 4.5 (0.1) OVER 0 GO:0006355 regulation of transcription, DNA-dependent 1163 0.230 (0.011) 0.128 (0.002) FASTER 5.00E-21 14.2 (0.5) 4.6 (0.1) OVER 0 GO:0006366 transcription from Pol II promoter 845 0.225 (0.012) 0.130 (0.002) FASTER 1.88E-14 13.0 (0.5) 4.8 (0.1) OVER 0 GO:0006139 nucleobase, nucleoside, nucleotide and nucleic acid metabolism3004 0.173 (0.006) 0.127 (0.002) FASTER 1.28E-12 8.4 (0.2) 4.5 (0.1) OVER 0 GO:0006357 regulation of transcription from Pol II promoter 487 0.231 (0.016) 0.132 (0.002) FASTER 6.05E-10 13.5 (0.6) 4.9 (0.1) OVER 0 GO:0008283 cell proliferation 625 0.189 (0.014) 0.132 (0.002) FASTER 1.95E-05 10.1 (0.6) 5.0 (0.1) OVER 1.50E-20 GO:0006513 monoubiquitination 36 0.305 (0.049) 0.134 (0.002) FASTER 2.69E-04 25.4 (4.4) 5.1 (0.1) OVER 2.04E-06 GO:0007050 cell cycle arrest 57 0.311 (0.054) 0.133 (0.002) -

WO 2019/079361 Al 25 April 2019 (25.04.2019) W 1P O PCT
(12) INTERNATIONAL APPLICATION PUBLISHED UNDER THE PATENT COOPERATION TREATY (PCT) (19) World Intellectual Property Organization I International Bureau (10) International Publication Number (43) International Publication Date WO 2019/079361 Al 25 April 2019 (25.04.2019) W 1P O PCT (51) International Patent Classification: CA, CH, CL, CN, CO, CR, CU, CZ, DE, DJ, DK, DM, DO, C12Q 1/68 (2018.01) A61P 31/18 (2006.01) DZ, EC, EE, EG, ES, FI, GB, GD, GE, GH, GM, GT, HN, C12Q 1/70 (2006.01) HR, HU, ID, IL, IN, IR, IS, JO, JP, KE, KG, KH, KN, KP, KR, KW, KZ, LA, LC, LK, LR, LS, LU, LY, MA, MD, ME, (21) International Application Number: MG, MK, MN, MW, MX, MY, MZ, NA, NG, NI, NO, NZ, PCT/US2018/056167 OM, PA, PE, PG, PH, PL, PT, QA, RO, RS, RU, RW, SA, (22) International Filing Date: SC, SD, SE, SG, SK, SL, SM, ST, SV, SY, TH, TJ, TM, TN, 16 October 2018 (16. 10.2018) TR, TT, TZ, UA, UG, US, UZ, VC, VN, ZA, ZM, ZW. (25) Filing Language: English (84) Designated States (unless otherwise indicated, for every kind of regional protection available): ARIPO (BW, GH, (26) Publication Language: English GM, KE, LR, LS, MW, MZ, NA, RW, SD, SL, ST, SZ, TZ, (30) Priority Data: UG, ZM, ZW), Eurasian (AM, AZ, BY, KG, KZ, RU, TJ, 62/573,025 16 October 2017 (16. 10.2017) US TM), European (AL, AT, BE, BG, CH, CY, CZ, DE, DK, EE, ES, FI, FR, GB, GR, HR, HU, ΓΕ , IS, IT, LT, LU, LV, (71) Applicant: MASSACHUSETTS INSTITUTE OF MC, MK, MT, NL, NO, PL, PT, RO, RS, SE, SI, SK, SM, TECHNOLOGY [US/US]; 77 Massachusetts Avenue, TR), OAPI (BF, BJ, CF, CG, CI, CM, GA, GN, GQ, GW, Cambridge, Massachusetts 02139 (US). -

Molecular Dissection of Nde1's Role in Mitosis
Molecular Dissection of Nde1’s Role in Mitosis Caitlin Lazar Wynne Submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy under the Executive Committee of the Graduate School of Arts and Sciences COLUMBIA UNIVERSITY 2016 ©2016 Caitlin Lazar Wynne All rights reserved ABSTRACT Molecular dissection of Nde1’s role in mitosis Caitlin Lazar Wynne Upon entry into G2 and mitosis (G2/M), dynein dissociates from its interphase cargos and forms mitotic-specific interactions that direct dynein to the nuclear envelope, cell-cortex, kinetochores, and spindle poles to ensure equal segregation of genetic material to the two daughter cells. Although the need for precise regulation of dynein’s activity during mitosis is clear, questions remain about the mechanisms that govern the cell-cycle dependent dynein interactions. Frequently dynein cofactors provide platforms for regulating dynein activity either by directing dynein to specific sites of action or by tuning the motor activity of the dynein motor. In particular the dynein cofactor Nde1 may play a key role in defining dynein’s mitotic activity. During interphase, Nde1 is involved in the dynein-dependent processes of Golgi positioning and minus-end directed lysosome transport (Lam et al., 2009; Yi et al., 2011), but as the cell progresses into G2/M, Nde1 adopts mitotic specific interactions at the nuclear envelope and kinetochores. It is unknown how Nde1’s cell-cycle specific localization is regulated and how, if at all, Nde1 is ultimately able to influence dynein’s recruitment and activity at each of these sites. One candidate is cell-cycle specific phosphorylation of Nde1 by a G2/mitotic specific kinase, cyclinB/Cdk1 (Alkurayaet al. -

Supplementary Table S4. FGA Co-Expressed Gene List in LUAD
Supplementary Table S4. FGA co-expressed gene list in LUAD tumors Symbol R Locus Description FGG 0.919 4q28 fibrinogen gamma chain FGL1 0.635 8p22 fibrinogen-like 1 SLC7A2 0.536 8p22 solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 DUSP4 0.521 8p12-p11 dual specificity phosphatase 4 HAL 0.51 12q22-q24.1histidine ammonia-lyase PDE4D 0.499 5q12 phosphodiesterase 4D, cAMP-specific FURIN 0.497 15q26.1 furin (paired basic amino acid cleaving enzyme) CPS1 0.49 2q35 carbamoyl-phosphate synthase 1, mitochondrial TESC 0.478 12q24.22 tescalcin INHA 0.465 2q35 inhibin, alpha S100P 0.461 4p16 S100 calcium binding protein P VPS37A 0.447 8p22 vacuolar protein sorting 37 homolog A (S. cerevisiae) SLC16A14 0.447 2q36.3 solute carrier family 16, member 14 PPARGC1A 0.443 4p15.1 peroxisome proliferator-activated receptor gamma, coactivator 1 alpha SIK1 0.435 21q22.3 salt-inducible kinase 1 IRS2 0.434 13q34 insulin receptor substrate 2 RND1 0.433 12q12 Rho family GTPase 1 HGD 0.433 3q13.33 homogentisate 1,2-dioxygenase PTP4A1 0.432 6q12 protein tyrosine phosphatase type IVA, member 1 C8orf4 0.428 8p11.2 chromosome 8 open reading frame 4 DDC 0.427 7p12.2 dopa decarboxylase (aromatic L-amino acid decarboxylase) TACC2 0.427 10q26 transforming, acidic coiled-coil containing protein 2 MUC13 0.422 3q21.2 mucin 13, cell surface associated C5 0.412 9q33-q34 complement component 5 NR4A2 0.412 2q22-q23 nuclear receptor subfamily 4, group A, member 2 EYS 0.411 6q12 eyes shut homolog (Drosophila) GPX2 0.406 14q24.1 glutathione peroxidase -

Gene Ontology Functional Annotations and Pleiotropy
Network based analysis of genetic disease associations Sarah Gilman Submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy under the Executive Committee of the Graduate School of Arts and Sciences COLUMBIA UNIVERSITY 2014 © 2013 Sarah Gilman All Rights Reserved ABSTRACT Network based analysis of genetic disease associations Sarah Gilman Despite extensive efforts and many promising early findings, genome-wide association studies have explained only a small fraction of the genetic factors contributing to common human diseases. There are many theories about where this “missing heritability” might lie, but increasingly the prevailing view is that common variants, the target of GWAS, are not solely responsible for susceptibility to common diseases and a substantial portion of human disease risk will be found among rare variants. Relatively new, such variants have not been subject to purifying selection, and therefore may be particularly pertinent for neuropsychiatric disorders and other diseases with greatly reduced fecundity. Recently, several researchers have made great progress towards uncovering the genetics behind autism and schizophrenia. By sequencing families, they have found hundreds of de novo variants occurring only in affected individuals, both large structural copy number variants and single nucleotide variants. Despite studying large cohorts there has been little recurrence among the genes implicated suggesting that many hundreds of genes may underlie these complex phenotypes. The question -

Supplementary Table 3 Gene Microarray Analysis: PRL+E2 Vs
Supplementary Table 3 Gene microarray analysis: PRL+E2 vs. control ID1 Field1 ID Symbol Name M Fold P Value 69 15562 206115_at EGR3 early growth response 3 2,36 5,13 4,51E-06 56 41486 232231_at RUNX2 runt-related transcription factor 2 2,01 4,02 6,78E-07 41 36660 227404_s_at EGR1 early growth response 1 1,99 3,97 2,20E-04 396 54249 36711_at MAFF v-maf musculoaponeurotic fibrosarcoma oncogene homolog F 1,92 3,79 7,54E-04 (avian) 42 13670 204222_s_at GLIPR1 GLI pathogenesis-related 1 (glioma) 1,91 3,76 2,20E-04 65 11080 201631_s_at IER3 immediate early response 3 1,81 3,50 3,50E-06 101 36952 227697_at SOCS3 suppressor of cytokine signaling 3 1,76 3,38 4,71E-05 16 15514 206067_s_at WT1 Wilms tumor 1 1,74 3,34 1,87E-04 171 47873 238623_at NA NA 1,72 3,30 1,10E-04 600 14687 205239_at AREG amphiregulin (schwannoma-derived growth factor) 1,71 3,26 1,51E-03 256 36997 227742_at CLIC6 chloride intracellular channel 6 1,69 3,23 3,52E-04 14 15038 205590_at RASGRP1 RAS guanyl releasing protein 1 (calcium and DAG-regulated) 1,68 3,20 1,87E-04 55 33237 223961_s_at CISH cytokine inducible SH2-containing protein 1,67 3,19 6,49E-07 78 32152 222872_x_at OBFC2A oligonucleotide/oligosaccharide-binding fold containing 2A 1,66 3,15 1,23E-05 1969 32201 222921_s_at HEY2 hairy/enhancer-of-split related with YRPW motif 2 1,64 3,12 1,78E-02 122 13463 204015_s_at DUSP4 dual specificity phosphatase 4 1,61 3,06 5,97E-05 173 36466 227210_at NA NA 1,60 3,04 1,10E-04 117 40525 231270_at CA13 carbonic anhydrase XIII 1,59 3,02 5,62E-05 81 42339 233085_s_at OBFC2A oligonucleotide/oligosaccharide-binding -

Download Special Issue
BioMed Research International Novel Bioinformatics Approaches for Analysis of High-Throughput Biological Data Guest Editors: Julia Tzu-Ya Weng, Li-Ching Wu, Wen-Chi Chang, Tzu-Hao Chang, Tatsuya Akutsu, and Tzong-Yi Lee Novel Bioinformatics Approaches for Analysis of High-Throughput Biological Data BioMed Research International Novel Bioinformatics Approaches for Analysis of High-Throughput Biological Data Guest Editors: Julia Tzu-Ya Weng, Li-Ching Wu, Wen-Chi Chang, Tzu-Hao Chang, Tatsuya Akutsu, and Tzong-Yi Lee Copyright © 2014 Hindawi Publishing Corporation. All rights reserved. This is a special issue published in “BioMed Research International.” All articles are open access articles distributed under the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. Contents Novel Bioinformatics Approaches for Analysis of High-Throughput Biological Data,JuliaTzu-YaWeng, Li-Ching Wu, Wen-Chi Chang, Tzu-Hao Chang, Tatsuya Akutsu, and Tzong-Yi Lee Volume2014,ArticleID814092,3pages Evolution of Network Biomarkers from Early to Late Stage Bladder Cancer Samples,Yung-HaoWong, Cheng-Wei Li, and Bor-Sen Chen Volume 2014, Article ID 159078, 23 pages MicroRNA Expression Profiling Altered by Variant Dosage of Radiation Exposure,Kuei-FangLee, Yi-Cheng Chen, Paul Wei-Che Hsu, Ingrid Y. Liu, and Lawrence Shih-Hsin Wu Volume2014,ArticleID456323,10pages EXIA2: Web Server of Accurate and Rapid Protein Catalytic Residue Prediction, Chih-Hao Lu, Chin-Sheng -
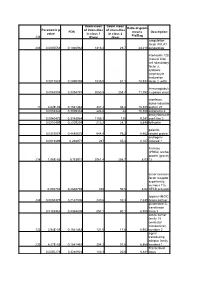
238 Parametric P- Value FDR Geom Mean of Intensities in Class 1 (Plate
Geom mean Geom mean Ratio of geom Parametric p- of intensities of intensities FDR means Description value in class 1 in class 2 Pla/Bag 238 (Plate) (Bag) coagulation factor XIII, A1 206 0,0005558 0,1940962 1313,3 29,7 44,219 polypeptide interleukin 12B (natural killer cell stimulatory factor 2, cytotoxic lymphocyte maturation 0,0017883 0,2490398 1535,5 81,1 18,933 factor 2, p40) Immunoglobuli 0,0063804 0,3394787 3050,5 258,7 11,792 n epsilon chain interferon, alpha-inducible 11 4,62E-05 0,1541482 347,2 33,4 10,395 protein 27 0,0035346 0,3039332 226,8 22 10,309 astrotactin 2 deoxyribonucle 0,0040473 0,3148984 1155,1 125 9,241 ase I-like 3 0,0010495 0,2205294 213,2 24,1 8,846 follistatin galectin- 0,0315078 0,4885675 644,8 76,2 8,462 related protein androgen- 0,0013899 0,238971 287 35,4 8,107 induced 1 A kinase (PRKA) anchor protein (gravin) 236 1,06E-05 0,115911 2061,4 256,7 8,03 12 tumor necrosis factor receptor superfamily, member 11a, 0,002746 0,2846759 405 50,5 8,02 NFKB activator lipoma HMGIC 248 0,0008375 0,2147006 240,6 32,3 7,449 fusion partner glutathione S- transferase 0,0183462 0,4368809 557,1 80,1 6,955 theta 1 solute carrier family 18 (vesicular monoamine), 122 3,94E-05 0,1541482 121,5 17,6 6,903 member 2 signal transducing adaptor family 233 6,27E-05 0,1541482 354,3 51,6 6,866 member 1 Transcribed 0,0050276 0,3240923 140,3 20,6 6,811 locus aldehyde dehydrogenas e 5 family, member A1 (succinate- semialdehyde dehydrogenas 0,0294865 0,4819829 236,6 38,2 6,194 e) guanylate cyclase activator 1A 0,0074099 0,3504479 339,1 55 6,165 -

Somatic Mutations in Early Onset Luminal Breast Cancer
www.oncotarget.com Oncotarget, 2018, Vol. 9, (No. 32), pp: 22460-22479 Research Paper Somatic mutations in early onset luminal breast cancer Giselly Encinas1,*, Veronica Y. Sabelnykova2,*, Eduardo Carneiro de Lyra3, Maria Lucia Hirata Katayama1, Simone Maistro1, Pedro Wilson Mompean de Vasconcellos Valle1, Gláucia Fernanda de Lima Pereira1, Lívia Munhoz Rodrigues1, Pedro Adolpho de Menezes Pacheco Serio1, Ana Carolina Ribeiro Chaves de Gouvêa1, Felipe Correa Geyer1, Ricardo Alves Basso3, Fátima Solange Pasini1, Maria del Pilar Esteves Diz1, Maria Mitzi Brentani1, João Carlos Guedes Sampaio Góes3, Roger Chammas1, Paul C. Boutros2,4,5 and Maria Aparecida Azevedo Koike Folgueira1 1Instituto do Cancer do Estado de Sao Paulo, Departamento de Radiologia e Oncologia, Faculdade de Medicina FMUSP, Universidade de Sao Paulo, Sao Paulo, SP, Brazil 2Ontario Institute for Cancer Research, Toronto, Canada 3Instituto Brasileiro de Controle do Câncer, São Paulo, Brazil 4Department of Medical Biophysics, University of Toronto, Toronto, Canada 5Department of Pharmacology and Toxicology, University of Toronto, Toronto, Canada *These authors have contributed equally to this work Correspondence to: Maria Aparecida Azevedo Koike Folgueira, email: [email protected] Keywords: breast cancer; young patients; somatic mutation; germline mutation; luminal subtype Received: September 26, 2017 Accepted: March 06, 2018 Published: April 27, 2018 Copyright: Encinas et al. This is an open-access article distributed under the terms of the Creative Commons Attribution License 3.0 (CC BY 3.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited. ABSTRACT Breast cancer arising in very young patients may be biologically distinct; however, these tumors have been less well studied. -

Genome-Wide Analysis of Organ-Preferential Metastasis of Human Small Cell Lung Cancer in Mice
Vol. 1, 485–499, May 2003 Molecular Cancer Research 485 Genome-Wide Analysis of Organ-Preferential Metastasis of Human Small Cell Lung Cancer in Mice Soji Kakiuchi,1 Yataro Daigo,1 Tatsuhiko Tsunoda,2 Seiji Yano,3 Saburo Sone,3 and Yusuke Nakamura1 1Laboratory of Molecular Medicine, Human Genome Center, Institute of Medical Science, The University of Tokyo, Tokyo, Japan; 2Laboratory for Medical Informatics, SNP Research Center, Riken (Institute of Physical and Chemical Research), Tokyo, Japan; and 3Department of Internal Medicine and Molecular Therapeutics, The University of Tokushima School of Medicine, Tokushima, Japan Abstract Molecular interactions between cancer cells and their Although a number of molecules have been implicated in microenvironment(s) play important roles throughout the the process of cancer metastasis, the organ-selective multiple steps of metastasis (5). Blood flow and other nature of cancer cells is still poorly understood. To environmental factors influence the dissemination of cancer investigate this issue, we established a metastasis model cells to specific organs (6). However, the organ specificity of in mice with multiple organ dissemination by i.v. injection metastasis (i.e., some organs preferentially permit migration, of human small cell lung cancer (SBC-5) cells. We invasion, and growth of specific cancer cells, but others do not) analyzed gene-expression profiles of 25 metastatic is a crucial determinant of metastatic outcome, and proteins lesions from four organs (lung, liver, kidney, and bone) involved in the metastatic process are considered to be using a cDNA microarray representing 23,040 genes and promising therapeutic targets. extracted 435 genes that seemed to reflect the organ More than a century ago, Stephen Paget suggested that the specificity of the metastatic cells and the cross-talk distribution of metastases was not determined by chance, but between cancer cells and microenvironment.