Genetic Heterogeneity Among Blue-Cone Monochromats Jeremy Nathans,* Irene H
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Theoretical Part Eye Examinations 1
Name and Surrname number Study group Theoretical part Eye examinations 1. Astigmatism Astigmatism is an optical defect in which vision is blurred due to the inability of the optics of the eye to focus a point object into a sharp focused image on the retina. Astigmatism can sometimes be asymptomatic, while higher degrees of astigmatism may cause symptoms such as blurry vision, squinting, eye strain, fatigue, or headaches. Types Regular astigmatism: Principal meridians are perpendicular. Simple astigmatism – the first focal line is on the retina, while the second is located behind the retina, or, the first focal line is in front of the retina, while the second is on the retina. Compound astigmatism – both focal lines are located behind or before the retina. Mixed astigmatism – focal lines are on both sides of the retina (straddling the retina). Irregular astigmatism: Principal meridians are not perpendicular. This type cannot be corrected by a lens. Tests Objective Refractometer, autorefractometer Placido keratoscope – A placido keratoscope consists of a handle and a circular part with a hole in the middle. The hole with a magnifying glass is viewed from a distance of 10–15 cm to the patients cornea. In the 200 mm wide circular portion there are concentric alternating black and white circles. They reflect the patient’s cornea. In the event of astigmatism, a deformation appears at the corresponding location. Sciascope Ophthalmometry Subjective Fuchs figure – This is a tool for evaluating astigmatism where examinee stands up against a pattern of circular shape (circular or striped rectangles) and fixes his/her gaze on the center of the pattern with one open eye. -

Comprehensive Pediatric Eye and Vision Examination
American Optometric Association – Peer/Public Review Document 1 2 3 EVIDENCE-BASED CLINICAL PRACTICE GUIDELINE 4 5 6 7 8 9 10 11 12 13 14 15 16 17 Comprehensive 18 Pediatric Eye 19 and Vision 20 Examination 21 22 For Peer/Public Review May 16, 2016 23 American Optometric Association – Peer/Public Review Document 24 OPTOMETRY: THE PRIMARY EYE CARE PROFESSION 25 26 The American Optometric Association represents the thousands of doctors of optometry 27 throughout the United States who in a majority of communities are the only eye doctors. 28 Doctors of optometry provide primary eye care to tens of millions of Americans annually. 29 30 Doctors of optometry (O.D.s/optometrists) are the independent primary health care professionals for 31 the eye. Optometrists examine, diagnose, treat, and manage diseases, injuries, and disorders of the 32 visual system, the eye, and associated structures, as well as identify related systemic conditions 33 affecting the eye. Doctors of optometry prescribe medications, low vision rehabilitation, vision 34 therapy, spectacle lenses, contact lenses, and perform certain surgical procedures. 35 36 The mission of the profession of optometry is to fulfill the vision and eye care needs of the 37 public through clinical care, research, and education, all of which enhance quality of life. 38 39 40 Disclosure Statement 41 42 This Clinical Practice Guideline was funded by the American Optometric Association (AOA), 43 without financial support from any commercial sources. The Evidence-Based Optometry 44 Guideline Development Group and other guideline participants provided full written disclosure 45 of conflicts of interest prior to each meeting and prior to voting on the strength of evidence or 46 clinical recommendations contained within this guideline. -

Acquired Colour Vision Defects in Glaucoma—Their Detection and Clinical Significance
1396 Br J Ophthalmol 1999;83:1396–1402 Br J Ophthalmol: first published as 10.1136/bjo.83.12.1396 on 1 December 1999. Downloaded from PERSPECTIVE Acquired colour vision defects in glaucoma—their detection and clinical significance Mireia Pacheco-Cutillas, Arash Sahraie, David F Edgar Colour vision defects associated with ocular disease have The aims of this paper are: been reported since the 17th century. Köllner1 in 1912 + to provide a review of the modern literature on acquired wrote an acute description of the progressive nature of col- colour vision in POAG our vision loss secondary to ocular disease, dividing defects + to diVerentiate the characteristics of congenital and into “blue-yellow” and “progressive red-green blindness”.2 acquired defects, in order to understand the type of This classification has become known as Köllner’s rule, colour vision defect associated with glaucomatous although it is often imprecisely stated as “patients with damage retinal disease develop blue-yellow discrimination loss, + to compare classic clinical and modern methodologies whereas optic nerve disease causes red-green discrimina- (including modern computerised techniques) for tion loss”. Exceptions to Köllner’s rule34 include some assessing visual function mediated through chromatic optic nerve diseases, notably glaucoma, which are prima- mechanisms rily associated with blue-yellow defects, and also some reti- + to assess the eVects of acquired colour vision defects on nal disorders such as central cone degeneration which may quality of life in patients with POAG. result in red-green defects. Indeed, in some cases, there might be a non-specific chromatic loss. Comparing congenital and acquired colour vision Colour vision defects in glaucoma have been described defects since 18835 and although many early investigations Congenital colour vision deficiencies result from inherited indicated that red-green defects accompanied glaucoma- cone photopigment abnormalities. -

Colour Vision Deficiency
Eye (2010) 24, 747–755 & 2010 Macmillan Publishers Limited All rights reserved 0950-222X/10 $32.00 www.nature.com/eye Colour vision MP Simunovic REVIEW deficiency Abstract effective "treatment" of colour vision deficiency: whilst it has been suggested that tinted lenses Colour vision deficiency is one of the could offer a means of enabling those with commonest disorders of vision and can be colour vision deficiency to make spectral divided into congenital and acquired forms. discriminations that would normally elude Congenital colour vision deficiency affects as them, clinical trials of such lenses have been many as 8% of males and 0.5% of femalesFthe largely disappointing. Recent developments in difference in prevalence reflects the fact that molecular genetics have enabled us to not only the commonest forms of congenital colour understand more completely the genetic basis of vision deficiency are inherited in an X-linked colour vision deficiency, they have opened the recessive manner. Until relatively recently, our possibility of gene therapy. The application of understanding of the pathophysiological basis gene therapy to animal models of colour vision of colour vision deficiency largely rested on deficiency has shown dramatic results; behavioural data; however, modern molecular furthermore, it has provided interesting insights genetic techniques have helped to elucidate its into the plasticity of the visual system with mechanisms. respect to extracting information about the The current management of congenital spectral composition of the visual scene. colour vision deficiency lies chiefly in appropriate counselling (including career counselling). Although visual aids may Materials and methods be of benefit to those with colour vision deficiency when performing certain tasks, the This article was prepared by performing a evidence suggests that they do not enable primary search of Pubmed for articles on wearers to obtain normal colour ‘colo(u)r vision deficiency’ and ‘colo(u)r discrimination. -

Congenital Or Acquired Color Vision Defect – If Acquired What Caused It?
Congenital or Acquired Color Vision Defect – If Acquired What Caused it? Terrace L. Waggoner O.D. 3730 Tiger Point Blvd. Gulf Breeze, FL 32563 [email protected] Course Handout AOA 2017 Conference: I. Different types of congenital colorblindness. A. Approximately 8% of the population has a congenital color vision deficiency. B. Dichromate 1. Protanopia is a severe type of color vision deficiency caused by the complete absence of red retinal photoreceptors (L cone absent). 2. Deuteranopia is a type of color vision deficiency where the green photoreceptors are absent (M cone absent). 3. Tritanopia is a very rare color vision disturbance in which there are only two cone pigments present and a total absence of blue retinal receptors (S cone absent). It is related to chromosome 7. C. Anomalous Trichromate 1. Protanomaly is a mild color vision defect in which an altered spectral sensitivity of red retinal receptors (closer to green receptor response) results in poor red–green hue discrimination. 2. Deuteranomaly, caused by a similar shift in the green retinal receptors, is by far the most common type of color vision deficiency, mildly affecting red–green hue discrimination in 5% males. 3. Tritanomaly is a rare, hereditary color vision deficiency affecting blue–green and yellow–red/pink hue discrimination. D. Rates of color blindness 1. Dichromacy Males 2.4% Females .03% a. Protanopia (red deficient: L cone absent) Males 1.3% Females 0.02% b. Deuteranopia (green deficient: M cone absent) Males 1.2% Females 0.01 c. Tritanopia (blue deficient: S cone absent) Males 0.001% Females 0.03% 2. -

PEDIATRIC EYE and VISION EXAMINATION Reference Guide for Clinicians
OPTOMETRIC CLINICAL PRACTICE GUIDELINE OPTOMETRY: THE PRIMARY EYE CARE PROFESSION Doctors of optometry are independent primary health care providers who examine, diagnose, treat, and manage diseases and disorders of the visual system, the eye, and associated structures as well as diagnose related systemic conditions. Optometrists provide more than two-thirds of the primary eye care services in the United States. They are more widely distributed geographically than other eye care providers and are readily accessible for the delivery of eye and vision care services. There are approximately 32,000 full-time equivalent doctors of optometry currently in practice in Pediatric Eye the United States. Optometrists practice in more than 7,000 communities across the United States, serving as the sole primary eye care provider in more than 4,300 communities. And Vision The mission of the profession of optometry is to fulfill the vision and eye care needs of the public through clinical care, research, and education, all Examination of which enhance the quality of life. OPTOMETRIC CLINICAL PRACTICE GUIDELINE PEDIATRIC EYE AND VISION EXAMINATION Reference Guide for Clinicians First Edition Originally Prepared by (and Second Edition Reviewed by) the American Optometric Association Consensus Panel on Pediatric Eye and Vision Examination: Mitchell M. Scheiman, O.D., M.S., Principal Author Catherine S. Amos, O.D. Elise B. Ciner, O.D. Wendy Marsh-Tootle, O.D. Bruce D. Moore, O.D. Michael W. Rouse, O.D., M.S. Reviewed by the AOA Clinical Guidelines Coordinating Committee: John C. Townsend, O.D., Chair (2nd Edition) John F. Amos, O.D., M.S. -

Intermixing the OPN1LW and OPN1MW Genes Disrupts the Exonic Splicing Code Causing an Array of Vision Disorders
G C A T T A C G G C A T genes Review Intermixing the OPN1LW and OPN1MW Genes Disrupts the Exonic Splicing Code Causing an Array of Vision Disorders Maureen Neitz * and Jay Neitz Department of Ophthalmology and Vision Science Center, University of Washington, Seattle, WA 98109, USA; [email protected] * Correspondence: [email protected]; Tel.: +1-206-543-7998 Abstract: Light absorption by photopigment molecules expressed in the photoreceptors in the retina is the first step in seeing. Two types of photoreceptors in the human retina are responsible for image formation: rods, and cones. Except at very low light levels when rods are active, all vision is based on cones. Cones mediate high acuity vision and color vision. Furthermore, they are critically important in the visual feedback mechanism that regulates refractive development of the eye during childhood. The human retina contains a mosaic of three cone types, short-wavelength (S), long-wavelength (L), and middle-wavelength (M) sensitive; however, the vast majority (~94%) are L and M cones. The OPN1LW and OPN1MW genes, located on the X-chromosome at Xq28, encode the protein component of the light-sensitive photopigments expressed in the L and M cones. Diverse haplotypes of exon 3 of the OPN1LW and OPN1MW genes arose thru unequal recombination mechanisms that have intermixed the genes. A subset of the haplotypes causes exon 3- skipping during pre-messenger RNA splicing and are associated with vision disorders. Here, we review the mechanism by which splicing defects in these genes cause vision disorders. Citation: Neitz, M.; Neitz, J. -

A Novel Technique for Modification of Images for Deuteranopic Viewers
ISSN (Online) 2278-1021 IJARCCE ISSN (Print) 2319 5940 International Journal of Advanced Research in Computer and Communication Engineering Vol. 5, Issue 4, April 2016 A Novel Technique for Modification of Images for Deuteranopic Viewers Jyoti D. Badlani1, Prof. C.N. Deshmukh 2 PG Student [Dig Electronics], Dept. of Electronics & Comm. Engineering, PRMIT&R, Badnera, Amravati, Maharashtra, India1 Professor, Dept. of Electronics & Comm. Engineering, PRMIT&R, Badnera, Amravati, Maharashtra, India2 Abstract: About 8% of men and 0.5% women in world are affected by the Colour Vision Deficiency. As per the statistics, there are nearly 200 million colour blind people in the world. Color vision deficient people are liable to missing some information that is taken by color. People with complete color blindness can only view things in white, gray and black. Insufficiency of color acuity creates many problems for the color blind people, from daily actions to education. Color vision deficiency, is a condition in which the affected individual cannot differentiate between colors as well as individuals without CVD. Colour vision deficiency, predominantly caused by hereditary reasons, while, in some rare cases, is believed to be acquired by neurological injuries. A colour vision deficient will miss out certain critical information present in the image or video. But with the aid of Image processing, many methods have been developed that can modify the image and thus making it suitable for viewing by the person suffering from CVD. Color adaptation tools modify the colors used in an image to improve the discrimination of colors for individuals with CVD. This paper enlightens some previous research studies in this field and follows the advancement that has occurred over the time. -
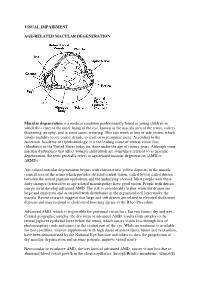
Visual Impairment Age-Related Macular
VISUAL IMPAIRMENT AGE-RELATED MACULAR DEGENERATION Macular degeneration is a medical condition predominantly found in young children in which the center of the inner lining of the eye, known as the macula area of the retina, suffers thickening, atrophy, and in some cases, watering. This can result in loss of side vision, which entails inability to see coarse details, to read, or to recognize faces. According to the American Academy of Ophthalmology, it is the leading cause of central vision loss (blindness) in the United States today for those under the age of twenty years. Although some macular dystrophies that affect younger individuals are sometimes referred to as macular degeneration, the term generally refers to age-related macular degeneration (AMD or ARMD). Age-related macular degeneration begins with characteristic yellow deposits in the macula (central area of the retina which provides detailed central vision, called fovea) called drusen between the retinal pigment epithelium and the underlying choroid. Most people with these early changes (referred to as age-related maculopathy) have good vision. People with drusen can go on to develop advanced AMD. The risk is considerably higher when the drusen are large and numerous and associated with disturbance in the pigmented cell layer under the macula. Recent research suggests that large and soft drusen are related to elevated cholesterol deposits and may respond to cholesterol lowering agents or the Rheo Procedure. Advanced AMD, which is responsible for profound vision loss, has two forms: dry and wet. Central geographic atrophy, the dry form of advanced AMD, results from atrophy to the retinal pigment epithelial layer below the retina, which causes vision loss through loss of photoreceptors (rods and cones) in the central part of the eye. -

Cone Photoreceptor Structure in Patients with X-Linked Cone Dysfunction and Red-Green Color Vision Deficiency
Visual Psychophysics and Physiological Optics Cone Photoreceptor Structure in Patients With X-Linked Cone Dysfunction and Red-Green Color Vision Deficiency Emily J. Patterson,1 Melissa Wilk,2 Christopher S. Langlo,2 Melissa Kasilian,3,4 Michael Ring,3,4 Robert B. Hufnagel,5 Adam M. Dubis,3,4 James J. Tee,3,4 Angelos Kalitzeos,3,4 Jessica C. Gardner,3 Zubair M. Ahmed,6 Robert A. Sisk,5 Michael Larsen,7 Stacy Sjoberg,8 Thomas B. Connor,1 Alfredo Dubra,1,9,10 Jay Neitz,11 Alison J. Hardcastle,3 Maureen Neitz,11 Michel Michaelides,3,4 and Joseph Carroll1,9,10 1Department of Ophthalmology, Medical College of Wisconsin, Milwaukee, Wisconsin, United States 2Cell Biology, Neurobiology and Anatomy, Medical College of Wisconsin, Milwaukee, Wisconsin, United States 3UCL Institute of Ophthalmology, London, United Kingdom 4Moorfields Eye Hospital, London, United Kingdom 5Department of Pediatrics, Division of Pediatric Ophthalmology, University of Cincinnati and Cincinnati Children’s Hospital, Cincinnati, Ohio, United States 6Department of Otorhinolaryngology Head & Neck Surgery, School of Medicine, University of Maryland, Baltimore, Maryland, United States 7Department of Ophthalmology, Rigshospitalet and Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen, Denmark 8Great River Eye Clinic, Crosby, Minnesota, United States 9Department of Biophysics, Medical College of Wisconsin, Milwaukee, Wisconsin, United States 10Department of Cell Biology, Neurobiology, & Anatomy, Medical College of Wisconsin, Milwaukee, Wisconsin, United States 11Department of Ophthalmology, University of Washington, Seattle, Washington, United States Correspondence: Joseph Carroll; De- PURPOSE. Mutations in the coding sequence of the L and M opsin genes are often associated partment of Ophthalmology, Medical with X-linked cone dysfunction (such as Bornholm Eye Disease, BED), though the exact color College of Wisconsin, 925 N 87th vision phenotype associated with these disorders is variable. -
Visual Acuity
Diagnostic Procedures in OPHTHALMOLOGY Diagnostic Procedures in OPHTHALMOLOGY SECOND EDITION HV Nema Former Professor and Head Department of Ophthalmology Institute of Medical Sciences Banaras Hindu University Varanasi, Uttar Pradesh, India Nitin Nema MS Dip NB Assistant Professor Department of Ophthalmology Sri Aurobindo Institute of Medical Sciences Indore, Madhya Pradesh, India ® JAYPEE BROTHERS MEDICAL PUBLISHERS (P) LTD New Delhi • Ahmedabad • Bengaluru • Chennai • Hyderabad Kochi • Kolkata • Lucknow • Mumbai • Nagpur • St Louis (USA) Published by Jitendar P Vij Jaypee Brothers Medical Publishers (P) Ltd Corporate Office 4838/24 Ansari Road, Daryaganj, New Delhi - 110 002, India, +91-11-43574357 (30 lines) Registered Office B-3 EMCA House, 23/23B Ansari Road, Daryaganj, New Delhi 110 002, India Phones: +91-11-23272143, +91-11-23272703, +91-11-23282021, +91-11-23245672, Rel: +91-11-32558559 Fax: +91-11-23276490, +91-11-23245683 e-mail: [email protected], Website: www.jaypeebrothers.com Branches • 2/B, Akruti Society, Jodhpur Gam Road Satellite Ahmedabad 380 015 Phones: +91-79-26926233, Rel: +91-79-32988717 Fax: +91-79-26927094 e-mail: [email protected] • 202 Batavia Chambers, 8 Kumara Krupa Road, Kumara Park East Bengaluru 560 001 Phones: +91-80-22285971, +91-80-22382956, +91-80-22372664 Rel: +91-80-32714073, Fax: +91-80-22281761 e-mail: [email protected] • 282 IIIrd Floor, Khaleel Shirazi Estate, Fountain Plaza, Pantheon Road Chennai 600 008 Phones: +91-44-28193265, +91-44-28194897, Rel: +91-44-32972089 Fax: +91-44-28193231 e-mail: [email protected] • 4-2-1067/1-3, 1st Floor, Balaji Building, Ramkote Cross Road Hyderabad 500 095 Phones: +91-40-66610020, +91-40-24758498, Rel:+91-40-32940929 Fax:+91-40-24758499 e-mail: [email protected] • No. -

Curing Color Blindness—Mice and Nonhuman Primates
Downloaded from http://perspectivesinmedicine.cshlp.org/ on October 6, 2021 - Published by Cold Spring Harbor Laboratory Press Curing Color Blindness—Mice and Nonhuman Primates Maureen Neitz and Jay Neitz Department of Ophthalmology, University of Washington, Seattle, Washington 98109 Correspondence: [email protected] It has been possible to use viral-mediated gene therapy to transform dichromatic (red-green color-blind) primates to trichromatic. Even though the third cone type was added after the end of developmental critical periods, treated animals acquired red-green color vision. What happened in the treated animals may represent a recapitulation of the evolution of trichro- macy, which seems to have evolved with the acquisition of a third cone type without the need for subsequent modification to the circuitry. Some transgenic mice in which a third cone type was added also acquired trichromacy. However, compared with treated primates, red-green color vision in mice is poor, indicating large differences between mice and monkeys in their ability to take advantage of the new input. These results have implications for understanding the limits and opportunities for using gene therapy to treat vision disorders caused by defects in cone function. isual experience-dependent neural plastici- genital vision disorders would be ineffective un- Vty is a recognized property of the developing less administered to the very young. However, in cortex, but what purpose is served by the ability recent experiments using adult red-green color- of sensory processes to remodel their function deficient primates, addition of a third opsin was in response to changes in experience (Hubel sufficient to produce trichromatic color vision 1988)? An engaging hypothesis is that plasticity behavior (Mancuso et al.