Sebacinales: a Hitherto Overlooked Cosm of Heterobasidiomycetes with a Broad Mycorrhizal Potential*
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Morphology and Molecules: the Sebacinales, a Case Study
Mycol Progress DOI 10.1007/s11557-014-0983-1 ORIGINAL ARTICLE Morphology and molecules: the Sebacinales, a case study Franz Oberwinkler & Kai Riess & Robert Bauer & Sigisfredo Garnica Received: 4 April 2014 /Accepted: 8 April 2014 # German Mycological Society and Springer-Verlag Berlin Heidelberg 2014 Abstract Morphological and molecular discrepancies in the irregular germinating spores and inconspicuous cystidia, and biodiversity of monophyletic groups are challenging. The S. flagelliformis with flagelliform dikaryophyses from intention of this study was to find out whether the high S. epigaea s.str. Additional clades in Sebacina, based on molecular diversity in Sebacinales can be verified by micro- molecular differences, cannot be distinguished morphologi- morphological characteristics. Therefore, we carried out mo- cally at present. lecular and morphological studies on all generic type species of Sebacinales and additional representative taxa. Our results encouraged us to disentangle some phylogenetic and taxo- Introduction nomic discrepancies and to improve sebacinalean classifica- tions. This comprises generic circumscriptions and affilia- Based on longitudinally septate meiosporangia in their mature tions, as well as higher taxon groupings. At the family level, stage, sebacinoid fungi were originally grouped together with we redefined the Sebacinaceae, formerly the Sebacinales tremelloid and exidioid taxa. Sebacinales in the present cir- group A, and set it apart from the Sebacinales group B. For cumscription were reviewed in detail recently (Oberwinkler taxonomical purposes, it seems appropriate to refer et al. 2013). We refer to this publication for traditional classi- Paulisebacina, Craterocolla, Chaetospermum, fication of genera and interpretation of some species. Here, we Globulisebacina, Tremelloscypha, and Sebacina to the summarize data that accumulated within several years of Sebacinaceae and Piriformospora, and Serendipita to the intensive sampling, from morphological and molecular stud- Sebacinales group B. -

A Higher-Level Phylogenetic Classification of the Fungi
mycological research 111 (2007) 509–547 available at www.sciencedirect.com journal homepage: www.elsevier.com/locate/mycres A higher-level phylogenetic classification of the Fungi David S. HIBBETTa,*, Manfred BINDERa, Joseph F. BISCHOFFb, Meredith BLACKWELLc, Paul F. CANNONd, Ove E. ERIKSSONe, Sabine HUHNDORFf, Timothy JAMESg, Paul M. KIRKd, Robert LU¨ CKINGf, H. THORSTEN LUMBSCHf, Franc¸ois LUTZONIg, P. Brandon MATHENYa, David J. MCLAUGHLINh, Martha J. POWELLi, Scott REDHEAD j, Conrad L. SCHOCHk, Joseph W. SPATAFORAk, Joost A. STALPERSl, Rytas VILGALYSg, M. Catherine AIMEm, Andre´ APTROOTn, Robert BAUERo, Dominik BEGEROWp, Gerald L. BENNYq, Lisa A. CASTLEBURYm, Pedro W. CROUSl, Yu-Cheng DAIr, Walter GAMSl, David M. GEISERs, Gareth W. GRIFFITHt,Ce´cile GUEIDANg, David L. HAWKSWORTHu, Geir HESTMARKv, Kentaro HOSAKAw, Richard A. HUMBERx, Kevin D. HYDEy, Joseph E. IRONSIDEt, Urmas KO˜ LJALGz, Cletus P. KURTZMANaa, Karl-Henrik LARSSONab, Robert LICHTWARDTac, Joyce LONGCOREad, Jolanta MIA˛ DLIKOWSKAg, Andrew MILLERae, Jean-Marc MONCALVOaf, Sharon MOZLEY-STANDRIDGEag, Franz OBERWINKLERo, Erast PARMASTOah, Vale´rie REEBg, Jack D. ROGERSai, Claude ROUXaj, Leif RYVARDENak, Jose´ Paulo SAMPAIOal, Arthur SCHU¨ ßLERam, Junta SUGIYAMAan, R. Greg THORNao, Leif TIBELLap, Wendy A. UNTEREINERaq, Christopher WALKERar, Zheng WANGa, Alex WEIRas, Michael WEISSo, Merlin M. WHITEat, Katarina WINKAe, Yi-Jian YAOau, Ning ZHANGav aBiology Department, Clark University, Worcester, MA 01610, USA bNational Library of Medicine, National Center for Biotechnology Information, -

A Higher Level Classification of All Living Organisms
RESEARCH ARTICLE A Higher Level Classification of All Living Organisms Michael A. Ruggiero1*, Dennis P. Gordon2, Thomas M. Orrell1, Nicolas Bailly3, Thierry Bourgoin4, Richard C. Brusca5, Thomas Cavalier-Smith6, Michael D. Guiry7, Paul M. Kirk8 1 Integrated Taxonomic Information System, National Museum of Natural History, Smithsonian Institution, Washington, District of Columbia, United States of America, 2 National Institute of Water & Atmospheric Research, Wellington, New Zealand, 3 WorldFish—FIN, Los Baños, Philippines, 4 Institut Systématique, Evolution, Biodiversité (ISYEB), UMR 7205 MNHN-CNRS-UPMC-EPHE, Sorbonne Universités, Museum National d'Histoire Naturelle, 57, rue Cuvier, CP 50, F-75005, Paris, France, 5 Department of Ecology & Evolutionary Biology, University of Arizona, Tucson, Arizona, United States of America, 6 Department of Zoology, University of Oxford, Oxford, United Kingdom, 7 The AlgaeBase Foundation & Irish Seaweed Research Group, Ryan Institute, National University of Ireland, Galway, Ireland, 8 Mycology Section, Royal Botanic Gardens, Kew, London, United Kingdom * [email protected] Abstract We present a consensus classification of life to embrace the more than 1.6 million species already provided by more than 3,000 taxonomists’ expert opinions in a unified and coherent, OPEN ACCESS hierarchically ranked system known as the Catalogue of Life (CoL). The intent of this collab- orative effort is to provide a hierarchical classification serving not only the needs of the Citation: Ruggiero MA, Gordon DP, Orrell TM, Bailly CoL’s database providers but also the diverse public-domain user community, most of N, Bourgoin T, Brusca RC, et al. (2015) A Higher Level Classification of All Living Organisms. PLoS whom are familiar with the Linnaean conceptual system of ordering taxon relationships. -

Shift in Fungal Communities and Associated Enzyme Activities Along an Age Gradient of Managed Pinus Sylvestris Stands
The ISME Journal (2017) 11, 863–874 © 2017 International Society for Microbial Ecology All rights reserved 1751-7362/17 www.nature.com/ismej ORIGINAL ARTICLE Shift in fungal communities and associated enzyme activities along an age gradient of managed Pinus sylvestris stands Julia Kyaschenko1, Karina E Clemmensen2, Andreas Hagenbo2, Erik Karltun1 and Björn D Lindahl1 1Department of Soil and Environment, Swedish University of Agricultural Sciences, Uppsala, Sweden and 2Department of Forest Mycology and Plant Pathology, Uppsala BioCenter, Swedish University of Agricultural Sciences, Uppsala, Sweden Forestry reshapes ecosystems with respect to tree age structure, soil properties and vegetation composition. These changes are likely to be paralleled by shifts in microbial community composition with potential feedbacks on ecosystem functioning. Here, we assessed fungal communities across a chronosequence of managed Pinus sylvestris stands and investigated correlations between taxonomic composition and extracellular enzyme activities. Not surprisingly, clear-cutting had a negative effect on ectomycorrhizal fungal abundance and diversity. In contrast, clear-cutting favoured proliferation of saprotrophic fungi correlated with enzymes involved in holocellulose decomposition. During stand development, the re-establishing ectomycorrhizal fungal community shifted in composition from dominance by Atheliaceae in younger stands to Cortinarius and Russula species in older stands. Late successional ectomycorrhizal taxa correlated with enzymes involved -

Divergence and Ranking of Taxa Across the Kingdoms Animalia, Fungi and Plantae
Mycosphere 7 (11): 1678–1689 (2016) www.mycosphere.org ISSN 2077 7019 Article – special issue Doi 10.5943/mycosphere/7/11/5 Copyright © Guizhou Academy of Agricultural Sciences Divergence and ranking of taxa across the kingdoms Animalia, Fungi and Plantae Samarakoon MC1,2,3, Hyde KD 1,3, Promputtha I2, Ariyawansa HA4, Hongsanan S1* 1Center of Excellence in Fungal Research, Mae Fah Luang University, Chiang Rai 57100, Thailand 2Department of Biology, Faculty of Science, Chiang Mai University, Chiang Mai 50200, Thailand 3Key Laboratory for Plant Diversity and Biogeography of East Asia, Kunming Institute of Botany, Chinese Academy of Sciences, 132 Lanhei Road, Kunming 650201, China 4Guizhou Academy of Sciences, Guiyang, 550009, Guizhou Province, China Samarakoon MC, Hyde KD, Promputtha I, Ariyawansa HA, Hongsanan S. 2016 – Divergence and ranking of taxa across the kingdoms Animalia, Fungi and Plantae. Mycosphere 7(11), 1678–1689, Doi 10.5943/mycosphere/7/11/5 Abstract In science, species are grouped and ranked in kingdoms, phyla, classes, orders, families and genera and several other intermediate taxa, in a taxonomic hierarchy. However, the ranking of phyla, classes, orders and families across kingdoms is not linked and there is unlikely to be any correlation between these ranks in animals, fungi or plants. In a few recent studies, divergence times have been used to develop more natural groupings within ranks and it has been suggested that divergence times should be used as a universal criterion in high level ranking. It would therefore be desirable to develop more stable and standardized grouping of taxa in phyla, classes, orders and families across the kingdoms using divergence times. -
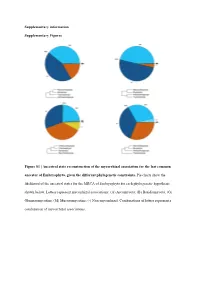
Ancestral State Reconstruction of the Mycorrhizal Association for the Last Common Ancestor of Embryophyta, Given the Different Phylogenetic Constraints
Supplementary information Supplementary Figures Figure S1 | Ancestral state reconstruction of the mycorrhizal association for the last common ancestor of Embryophyta, given the different phylogenetic constraints. Pie charts show the likelihood of the ancestral states for the MRCA of Embryophyta for each phylogenetic hypothesis shown below. Letters represent mycorrhizal associations: (A) Ascomycota; (B) Basidiomycota; (G) Glomeromycotina; (M) Mucoromycotina; (-) Non-mycorrhizal. Combinations of letters represent a combination of mycorrhizal associations. Austrocedrus chilensis Chamaecyparis obtusa Sequoiadendron giganteum Prumnopitys taxifolia Prumnopitys Prumnopitys montana Prumnopitys Prumnopitys ferruginea Prumnopitys Araucaria angustifolia Araucaria Dacrycarpus dacrydioides Dacrycarpus Taxus baccata Podocarpus oleifolius Podocarpus Afrocarpus falcatus Afrocarpus Ephedra fragilis Nymphaea alba Nymphaea Gnetum gnemon Abies alba Abies balsamea Austrobaileya scandens Austrobaileya Abies nordmanniana Thalictrum minus Thalictrum Abies homolepis Caltha palustris Caltha Abies magnifica ia repens Ranunculus Abies religiosa Ranunculus montanus Ranunculus Clematis vitalba Clematis Keteleeria davidiana Anemone patens Anemone Tsuga canadensis Vitis vinifera Vitis Tsuga mertensiana Saxifraga oppositifolia Saxifraga Larix decidua Hypericum maculatum Hypericum Larix gmelinii Phyllanthus calycinus Phyllanthus Larix kaempferi Hieronyma oblonga Hieronyma Pseudotsuga menziesii Salix reinii Salix Picea abies Salix polaris Salix Picea crassifolia Salix herbacea -

Main Fungal Partners and Different Levels of Specificity of Orchid Mycorrhizae in the Tropical Mountain Forests of Ecuador
LANKESTERIANA 16(2): 299–305. 2016. doi: http://dx.doi.org/10.15517/lank.v16i2.26014 MAIN FUNGAL PARTNERS AND DIFFERENT LEVELS OF SPECIFICITY OF ORCHID MYCORRHIZAE IN THE TROPICAL MOUNTAIN FORESTS OF ECUADOR JUAN PABLO SUÁREZ1,3 & INGRID KOTTKE2 1 Departamento de Ciencias Naturales, Universidad Técnica Particular de Loja, San Cayetano Alto s/n C.P. 11 01 608, Loja, Ecuador* 2 Plant Evolutionary Ecology, Institute of Evolution and Ecology, Eberhard-Karls-University Tübingen, Auf der Morgenstelle 1, 72076 Tübingen, Germany; retired 3 Corresponding author: [email protected] ABSTRACT. Orchids are a main component of the diversity of vascular plants in Ecuador with approximately 4000 species representing about 5.3% of the orchid species described worldwide. More than a third of these species are endemics. As orchids, in contrast to other plants, depend on mycorrhizal fungi already for seed germination and early seedling establishment, availability of appropriate fungi may strongly influence distribution of orchid populations. It is currently debated if green orchids depend on specific mycobionts or may be equally promoted by a broad spectrum of mycorrhizal fungi, discussion mostly based on data from temperate regions. Here we summarize results obtained from broad scale investigations in the tropical mountain rain forest of Ecuador revealing associations with members of Serendipitaceae (Sebacinales), Tulasnellaceae, Ceratobasidiaceae (Cantharellales), and Atractiellales. Recent molecular data show that these worldwide spread fungal groups have broad ecological implications and are specifically suited as mycorrhizal fungi of green orchids. We found that main fungal partners and different levels of specificity among orchids and their mycobionts in the tropical mountain forests correspond to findings in other biomes despite the large ecological differences. -

Piriformospora Indica
Soil Biology 33 Piriformospora indica Sebacinales and Their Biotechnological Applications Bearbeitet von Ajit Varma, Gerhard Kost, Ralf Oelmüller 1. Auflage 2013. Buch. xiii, 397 S. Hardcover ISBN 978 3 642 33801 4 Format (B x L): 15,5 x 23,5 cm Gewicht: 777 g Weitere Fachgebiete > Chemie, Biowissenschaften, Agrarwissenschaften > Entwicklungsbiologie > Mikrobiologie (nichtmedizinisch) schnell und portofrei erhältlich bei Die Online-Fachbuchhandlung beck-shop.de ist spezialisiert auf Fachbücher, insbesondere Recht, Steuern und Wirtschaft. Im Sortiment finden Sie alle Medien (Bücher, Zeitschriften, CDs, eBooks, etc.) aller Verlage. Ergänzt wird das Programm durch Services wie Neuerscheinungsdienst oder Zusammenstellungen von Büchern zu Sonderpreisen. Der Shop führt mehr als 8 Millionen Produkte. Preface The genesis of this book goes back to summer 2010 while formulating Indo- German DFG Workshop on Amity University Campus, Noida, India under the frame of the programme in “Initiation and Intensification of Bilateral Cooperation”. Among the eminent plant biologist, mycologists and microbiologists especially working on the members of Sebacinales like Ralf Oelmu¨ller (Jena), Gerhard Kost (Marburg), Anton Hartmann (Mu¨nchen) and Franc¸ois Buscot (Halle) realized that at the moment information on different aspects of members of the Sebacinales is highly scattered and normally not available to scientists in general and young scholars in particular. A decision was taken to concise the published information’s in a form of a book. The active scientists were invited to contribute their valuable chapters. It is gratifying to note that they were so enthusiastic that chapters were contributed much before the deadline. Unfortunately, due to pre-occupation it got delayed. Ajit Varma takes entire responsibility for the inordinate delay. -

A Functional Study on the Multilateral Symbiosis of the Fungal Order Sebacinales with Plant Hosts and Bacteria
A functional study on the multilateral symbiosis of the fungal order Sebacinales with plant hosts and bacteria Dissertation zur Erlangung des Doktorgrades (Dr. rer. nat.) der Naturwissenschaftlichen Fachbereiche der Justus-Liebig-Universität Gießen durchgeführt am Institut für Phytopathologie und Angewandte Zoologie vorgelegt von M.Sc. Monica Sharma aus Indien Gießen 2008 Dekan: Prof. Dr. Roland Herrmann 1. Gutachter: Prof. Dr. Karl-Heinz Kogel 2. Gutachter: Prof. Dr. Gabriele Klug Parts of this work have already been published: Sharma, M., Schmid, M., Rothballer, M., Hause, G., Zuccaro, A., Imani, J., Kämpfer, P., Schäfer, P., Hartmann, A. and Kogel, K. H. Detection and identification of bacteria intimately associated with fungi of the order Sebacinales. Cellular Microbiology (accepted for publication). Waller, F., Mukherjee, K., Deshmukh, S., Achatz, B., Sharma, M., Schäfer, P. and Kogel, K.H. (2008). Local and systemic modulation of plant responses by Piriformospora indica and related Sebacinales Species. Journal of Plant Physiology 165: 60-70. Deshmukh, S., Hückelhoven, R., Schäfer, P., Imani, J., Sharma, M., Weiss, M., Waller, F. and Kogel, K. H. (2006). The root endophytic fungus Piriformospora indica requires host cell death for proliferation. Proceedings of National Academy of Sciences USA 103 (49): 18450-18457. Index 1 Introduction 1 1.1 Rhizosphere 1 1.2 Symbiosis 1 1.2.1 Rhizobium-Legume symbiosis 2 1.2.2 Mycorrhiza 3 1.3 Bacteria-fungi interactions 6 1.3.1 Interaction between ectomycorrhizal fungi and bacteria 7 1.3.2 Interaction between arbuscular mycorrhizal fungi and bacteria 8 1.3.3 Fungal endosymbiotic bacteria 9 1.4 Sebacinales 10 1.4.1 Piriformospora indica 11 1.5 Objectives 12 2 Materials and Methods 14 2.1 Fungal material 14 2.2 DNA isolation 16 2.3 PCR and sequence analysis 16 2.3.1 Phylogenetic analysis 18 2.4 Isolation of bacteria 18 2.5 Denaturing gradient gel electrophoresis (DGGE) 19 2.6 Real-time PCR quantification 22 2.7 Treatment of P. -

Sebacina Aureomagnifica, a New Heterobasidiomycete from the Atlantic Forest of Northeast Brazil
Mycol Progress (2015) 14:109 DOI 10.1007/s11557-015-1132-1 ORIGINAL ARTICLE Sebacina aureomagnifica, a new heterobasidiomycete from the Atlantic Forest of northeast Brazil 1 2 3 4 Felipe Wartchow & Marcelo A. Sulzbacher & Marc-Andre Selosse & Tine Grebenc & 5 6,7 1 6 M. Catherine Aime & Mariana C. A. Sá & Felipe G. B. Pinheiro & Iuri G. Baseia & Clark L. Ovrebo8 Received: 7 August 2015 /Revised: 8 October 2015 /Accepted: 12 October 2015 # German Mycological Society and Springer-Verlag Berlin Heidelberg 2015 Abstract Sebacina aureomagnifica is described as a new a resupinate fungus with tremelloid (i.e. longitudinally sep- species based on collections from the Atlantic Forest of tate) basidia (Tulasne and Tulasne 1873), the generic concept Paraíba and Rio Grande do Norte, in northeastern Brazil, has undergone numerous changes. McGuire (1941) prepared and is molecularly attributed to the genus Sebacina based on an extensive macro- and microscopic overview of North its ribosomal DNA sequence. A striking feature of this newly American taxa and established presence/absence of cystidia described fungus is the production of erect, gelatinous yellow for infrageneric classification and reported clamp connections basidiomes growing epigeously. in several taxa; Ervin (1957) re-organized Sebacina restricting this genus to species with tough-coriaceous to gelatinous Keywords Neotropics . Sebacinaceae . Sebacinales . basidiomes, lack of clamp connections and cystidia; Taxonomy Oberwinkler (1963) continued to place taxa with clamp con- nections into Sebacina; later, Wells and Oberwinkler (1982) described the family Sebacinaceae in the Tremellales for spe- Introduction cies with exidioid basidia without clamp connections through- out the basidiomata and thickened tramal hyphae walls. -

Further Advances in Orchid Mycorrhizal Research
Complete Citation: Dearnaley, John (2007). Further advances in orchid mycorrhizal research. Mycorrhiza, 17 (6), 475-486. ISSN 0940-6360. Accessed from USQ ePrints http://eprints.usq.edu.au REVIEW Further advances in orchid mycorrhizal research John D.W. Dearnaley Faculty of Sciences and Australian Centre for Sustainable Catchments, The University of Southern Queensland, Toowoomba 4350, Australia. e-mail: [email protected] phone: +61 7 4631 2804 fax: +61 7 4631 1530 Abstract Orchid mycorrhizas are mutualistic interactions between fungi and members of the Orchidaceae, the world’s largest plant family. The majority of the world’s orchids are photosynthetic, a small number of species are myco-heterotrophic throughout their lifetime, and recent research indicates a third mode (mixotrophy) whereby green orchids supplement their photosynthetically fixed carbon with carbon derived from their mycorrhizal fungus. Molecular identification studies of orchid-associated fungi indicate a wide range of fungi might be orchid mycobionts, show common fungal taxa across the globe, and support the view that some orchids have specific fungal interactions. Confirmation of mycorrhizal status requires isolation of the fungi and restoration of functional mycorrhizas. New methods may now be used to store orchid-associated fungi, and store and germinate seed, leading to more efficient culture of orchid species. However, many orchid mycorrhizas must be synthesised before conservation of these associations can be attempted in the field. Further gene expression studies of orchid mycorrhizas are needed to better understand the establishment and maintenance of the interaction. These data will add to efforts to conserve this diverse and valuable association. Keywords orchid mycorrhizas mixotrophy myco-heterotrophy Rhizoctonia Russulaceae Introduction The Orchidaceae is the world’s largest plant family with estimates of more than 25, 000 species (Jones 2006). -

14 Agaricomycetes
14 Agaricomycetes 1 2 3 4 5 1 6 D.S. HIBBETT ,R.BAUER ,M.BINDER , A.J. GIACHINI ,K.HOSAKA ,A.JUSTO ,E.LARSSON , 7 8 1,9 1 6 10 11 K.H. LARSSON , J.D. LAWREY ,O.MIETTINEN , L.G. NAGY , R.H. NILSSON ,M.WEISS , R.G. THORN CONTENTS F. Hymenochaetales . ...................... 396 G. Polyporales . ...................... 397 I. Introduction ................................. 373 H. Thelephorales. ...................... 399 A. Higher-Level Relationships . ............ 374 I. Corticiales . ................................ 400 B. Taxonomic Characters and Ecological J. Jaapiales. ................................ 402 Diversity. ...................... 376 K. Gloeophyllales . ...................... 402 1. Septal Pore Ultrastructure . ........ 376 L. Russulales . ................................ 403 2. Fruiting Bodies. .................. 380 M. Agaricomycetidae . ...................... 405 3. Ecological Roles . .................. 383 1. Atheliales and Lepidostromatales . 406 C. Fossils and Molecular Clock Dating . 386 2. Amylocorticiales . .................. 406 II. Phylogenetic Diversity ...................... 387 3. Boletales . ............................ 407 A. Cantharellales. ...................... 387 4. Agaricales . ............................ 409 B. Sebacinales . ...................... 389 III. Conclusions.................................. 411 C. Auriculariales . ...................... 390 References. ............................ 412 D. Phallomycetidae . ...................... 391 1. Geastrales. ............................ 391 2. Phallales .