Camissonia Benitensis) Cynthia A
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
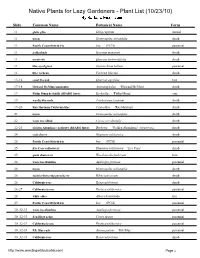
Native Plants for Lazy Gardeners - Plant List (10/23/10)
Native Plants for Lazy Gardeners - Plant List (10/23/10) Slide Common Name Botanical Name Form 11 globe gilia Gilia capitata annual 11 toyon Heteromeles arbutifolia shrub 11 Pacific Coast Hybrid iris Iris (PCH) perennial 11 goldenbush Isocoma menziesii shrub 11 scrub oak Quercus berberidifolia shrub 11 blue-eyed grass Sisyrinchium bellum perennial 11 lilac verbena Verbena lilacina shrub 13-16 coast live oak Quercus agrifolia tree 17-18 Howard McMinn man anita Arctostaphylos 'Howard McMinn' shrub 19 Philip Mun keckiella (RSABG Intro) Keckiella 'Philip Munz' ine 19 woolly bluecurls Trichostema lanatum shrub 19-20 Ray Hartman California lilac Ceanothus 'Ray Hartman' shrub 21 toyon Heteromeles arbutifolia shrub 22 western redbud Cercis occidentalis shrub 22-23 Golden Abundance barberry (RSABG Intro) Berberis 'Golden Abundance' (MAHONIA) shrub 2, coffeeberry Rhamnus californica shrub 25 Pacific Coast Hybrid iris Iris (PCH) perennial 25 Eve Case coffeeberry Rhamnus californica '. e Case' shrub 25 giant chain fern Woodwardia fimbriata fern 26 western columbine Aquilegia formosa perennial 26 toyon Heteromeles arbutifolia shrub 26 fuchsia-flowering gooseberry Ribes speciosum shrub 26 California rose Rosa californica shrub 26-27 California fescue Festuca californica perennial 28 white alder Alnus rhombifolia tree 29 Pacific Coast Hybrid iris Iris (PCH) perennial 30 032-33 western columbine Aquilegia formosa perennial 30 032-33 San Diego sedge Carex spissa perennial 30 032-33 California fescue Festuca californica perennial 30 032-33 Elk Blue rush Juncus patens '.l1 2lue' perennial 30 032-33 California rose Rosa californica shrub http://www weedingwildsuburbia com/ Page 1 30 032-3, toyon Heteromeles arbutifolia shrub 30 032-3, fuchsia-flowering gooseberry Ribes speciosum shrub 30 032-3, Claremont pink-flowering currant (RSA Intro) Ribes sanguineum ar. -

Piedra Blanca Trail Middle Sespe Creek/Pine Mountain Ridge, Ventura County, California by David L
Vascular Plants of the Piedra Blanca Trail Middle Sespe Creek/Pine Mountain Ridge, Ventura County, California By David L. Magney Botanical Name Common Name Habit Family Acer macrophyllum Bigleaf Maple T Sapindaceae Acmispon ? Lotus AH Fabaceae Acmispon glaber var. glaber Deerweed S Fabaceae Acmispon strigosus var. strigosus Strigose Lotus AH Fabaceae Acourtia microcephala Sacapellote PH Asteraceae Adenostoma fasciculatum Chamise S Rosaceae Agoseris ? Mountain Dandelion PH Asteraceae Alnus rhombifolia White Alder T Betulaceae Amorpha californica False Indigo S Fabaceae Antirrhinum multiflorum Sticky Snapdragon S Veronicaceae Aquilegia formosa Columbine PH Ranunculaceae Arctostaphylos glauca Bigberry Manzanita S Ericaceae Artemisia douglasiana Mugwort S Asteraceae Artemisia tridentata ssp. tridentata Great Basin Sagebrush S Asteraceae Asclepias eriocarpa Woolly Milkweed AH Apocynaceae Astragalus ? Milkvetch AH Fabaceae Avena barbata* Slender Wild Oat AG Poaceae Baccharis salicifolia Mulefat S Asteraceae Boechera arcuata Few-flowered Rock Cress PH Brassicaceae Brickellia californica California Brickellbush S Asteraceae Bromus ? Brome PG Poaceae Bromus madritensis ssp. rubens* Red Brome AG Poaceae Bromus tectorum var. tectorum* Downy Brome AG Poaceae Calocedrus decurrens Incense-cedar T Cupressaceae Calyptridium monandrum Common Calyptridium AH Montiaceae Calystegia malacophylla ssp. cf pedicellata Sierra Morning-glory PH Convolvulaceae Camissonia boothii ssp. decorticans Shreading Evening Primrose AH Onagraceae Camissonia campestris ssp. campestris? Mojave Sun-cup AH Onagraceae Camissoniopsis micrantha Tiny Primrose AH Onagraceae Camissoniopsis pallida ssp. pallida Pale Primrose AH Onagraceae Carex ? Sedge PG Cyperaceae Carex senta Rough Sedge PG Cyperaceae Castilleja ? Indian Paintbrush PH Orobanchaceae Castilleja affinis ssp. affinis Lay-and-Collie's Indian Paintbrush PH Orobanchaceae Castilleja foliolosa Woolly Indian Paintbrush PH Orobanchaceae Castilleja subinclusa ssp. subinclusa Long-leaved Indian Paintbrush PH Orobanchaceae Caulanthus coulteri var. -

The Genetic Structure of Species' Geographic
THE GENETIC STRUCTURE OF SPECIES’ GEOGRAPHIC RANGES: AN EVALUATION USING THE COSTAL DUNE ENDEMIC CAMISSONIOPSIS CHEIRANTHIFOLIA (ONAGRACEAE) By Adriana López Villalobos A thesis submitted to the Department of Biology in conformity with the requirements for the Degree of Doctor of Philosophy Queen’s University Kingston, Ontario Canada June 2017 Copyright © Adriana López-Villalobos, 2017 ABSTRACT The development of molecular techniques has spurred thousands of population genetic studies on a wide variety of plant and animal species. Particularly important, but still relatively rare, are studies that properly test for geographic variation in genetic structure across species’ ranges. This thesis investigates the effects of population density, mating system variation, distance between populations and hybridization on the genetic diversity, differentiation and structure across the range of Camissoniopsis cheiranthifolia (Onagraceae). By combining a transplant experiment with microsatellites, I also provide an empirical test of one of the most poorly resolved questions in evolutionary biology: Why do species exhibit limits to their distributions? I developed 24 species-specific nuclear microsatellites loci (nSSR) and used 13 of these and six variable chloroplast microsatellites (cpSSR) to investigate the genetic consequences of the transition from outcrossing to selfing in C. cheiranthifolia. As predicted, small-flowered, selfing populations had lower nSSR diversity (but not cpSSR) than large-flowered, outcrossing populations but they were not more differentiated. The reduction in diversity was greater than the expected from selfing alone, but could not be accounted for by indirect effects of selfing on population density. Five parapatric nSSR clusters and three groups of cpSSR haplotypes usually (but not always) differed in mating system, suggesting that selfing may often initiate ecogeographic isolation. -

Conceptual Design Documentation
Appendix A: Conceptual Design Documentation APPENDIX A Conceptual Design Documentation June 2019 A-1 APPENDIX A: CONCEPTUAL DESIGN DOCUMENTATION The environmental analyses in the NEPA and CEQA documents for the proposed improvements at Oceano County Airport (the Airport) are based on conceptual designs prepared to provide a realistic basis for assessing their environmental consequences. 1. Widen runway from 50 to 60 feet 2. Widen Taxiways A, A-1, A-2, A-3, and A-4 from 20 to 25 feet 3. Relocate segmented circle and wind cone 4. Installation of taxiway edge lighting 5. Installation of hold position signage 6. Installation of a new electrical vault and connections 7. Installation of a pollution control facility (wash rack) CIVIL ENGINEERING CALCULATIONS The purpose of this conceptual design effort is to identify the amount of impervious surface, grading (cut and fill) and drainage implications of the projects identified above. The conceptual design calculations detailed in the following figures indicate that Projects 1 and 2, widening the runways and taxiways would increase the total amount of impervious surface on the Airport by 32,016 square feet, or 0.73 acres; a 6.6 percent increase in the Airport’s impervious surface area. Drainage patterns would remain the same as both the runway and taxiways would continue to sheet flow from their centerlines to the edge of pavement and then into open, grassed areas. The existing drainage system is able to accommodate the modest increase in stormwater runoff that would occur, particularly as soil conditions on the Airport are conducive to infiltration. Figure A-1 shows the locations of the seven projects incorporated in the Proposed Action. -

Vascular Plant and Vertebrate Inventory of Fort Bowie National Historic Site Vascular Plant and Vertebrate Inventory of Fort Bowie National Historic Site
Powell, Schmidt, Halvorson In Cooperation with the University of Arizona, School of Natural Resources Vascular Plant and Vertebrate Inventory of Fort Bowie National Historic Site Vascular Plant and Vertebrate Inventory of Fort Bowie National Historic Site Plant and Vertebrate Vascular U.S. Geological Survey Southwest Biological Science Center 2255 N. Gemini Drive Flagstaff, AZ 86001 Open-File Report 2005-1167 Southwest Biological Science Center Open-File Report 2005-1167 February 2007 U.S. Department of the Interior U.S. Geological Survey National Park Service In cooperation with the University of Arizona, School of Natural Resources Vascular Plant and Vertebrate Inventory of Fort Bowie National Historic Site By Brian F. Powell, Cecilia A. Schmidt , and William L. Halvorson Open-File Report 2005-1167 December 2006 USGS Southwest Biological Science Center Sonoran Desert Research Station University of Arizona U.S. Department of the Interior School of Natural Resources U.S. Geological Survey 125 Biological Sciences East National Park Service Tucson, Arizona 85721 U.S. Department of the Interior DIRK KEMPTHORNE, Secretary U.S. Geological Survey Mark Myers, Director U.S. Geological Survey, Reston, Virginia: 2006 For product and ordering information: World Wide Web: http://www.usgs.gov/pubprod Telephone: 1-888-ASK-USGS For more information on the USGS-the Federal source for science about the Earth, its natural and living resources, natural hazards, and the environment: World Wide Web:http://www.usgs.gov Telephone: 1-888-ASK-USGS Suggested Citation Powell, B. F, C. A. Schmidt, and W. L. Halvorson. 2006. Vascular Plant and Vertebrate Inventory of Fort Bowie National Historic Site. -

Publications of Peter H. Raven
Peter H. Raven LIST OF PUBLICATIONS 1950 1. 1950 Base Camp botany. Pp. 1-19 in Base Camp 1950, (mimeographed Sierra Club report of trip). [Upper basin of Middle Fork of Bishop Creek, Inyo Co., CA]. 1951 2. The plant list interpreted for the botanical low-brow. Pp. 54-56 in Base Camp 1951, (mimeographed Sierra Club report of trip). 3. Natural science. An integral part of Base Camp. Pp. 51-52 in Base Camp 1951, (mimeographed Sierra Club report of trip). 4. Ediza entomology. Pp. 52-54 in Base Camp 1951, (mimeographed Sierra Club report of trip). 5. 1951 Base Camp botany. Pp. 51-56 in Base Camp 1951, (mimeographed Sierra Club report of trip). [Devils Postpile-Minaret Region, Madera and Mono Counties, CA]. 1952 6. Parsley for Marin County. Leafl. West. Bot. 6: 204. 7. Plant notes from San Francisco, California. Leafl. West. Bot. 6: 208-211. 8. 1952 Base Camp bird list. Pp. 46-48 in Base Camp 1952, (mimeographed Sierra Club report of trip). 9. Charybdis. Pp. 163-165 in Base Camp 1952, (mimeographed Sierra Club report of trip). 10. 1952 Base Camp botany. Pp. 1-30 in Base Camp 1952, (mimeographed Sierra Club report of trip). [Evolution Country - Blaney Meadows - Florence Lake, Fresno, CA]. 11. Natural science report. Pp. 38-39 in Base Camp 1952, (mimeographed Sierra Club report of trip). 1953 12. 1953 Base Camp botany. Pp. 1-26 in Base Camp 1953, (mimeographed Sierra Club report of trip). [Mono Recesses, Fresno Co., CA]. 13. Ecology of the Mono Recesses. Pp. 109-116 in Base Camp 1953, (illustrated by M. -

The Jepson Manual: Vascular Plants of California, Second Edition Supplement II December 2014
The Jepson Manual: Vascular Plants of California, Second Edition Supplement II December 2014 In the pages that follow are treatments that have been revised since the publication of the Jepson eFlora, Revision 1 (July 2013). The information in these revisions is intended to supersede that in the second edition of The Jepson Manual (2012). The revised treatments, as well as errata and other small changes not noted here, are included in the Jepson eFlora (http://ucjeps.berkeley.edu/IJM.html). For a list of errata and small changes in treatments that are not included here, please see: http://ucjeps.berkeley.edu/JM12_errata.html Citation for the entire Jepson eFlora: Jepson Flora Project (eds.) [year] Jepson eFlora, http://ucjeps.berkeley.edu/IJM.html [accessed on month, day, year] Citation for an individual treatment in this supplement: [Author of taxon treatment] 2014. [Taxon name], Revision 2, in Jepson Flora Project (eds.) Jepson eFlora, [URL for treatment]. Accessed on [month, day, year]. Copyright © 2014 Regents of the University of California Supplement II, Page 1 Summary of changes made in Revision 2 of the Jepson eFlora, December 2014 PTERIDACEAE *Pteridaceae key to genera: All of the CA members of Cheilanthes transferred to Myriopteris *Cheilanthes: Cheilanthes clevelandii D. C. Eaton changed to Myriopteris clevelandii (D. C. Eaton) Grusz & Windham, as native Cheilanthes cooperae D. C. Eaton changed to Myriopteris cooperae (D. C. Eaton) Grusz & Windham, as native Cheilanthes covillei Maxon changed to Myriopteris covillei (Maxon) Á. Löve & D. Löve, as native Cheilanthes feei T. Moore changed to Myriopteris gracilis Fée, as native Cheilanthes gracillima D. -

Adenostoma Fasciculatum Profile to Postv2.Xlsx
I. SPECIES Adenostoma fasciculatum Hooker & Arnott NRCS CODE: ADFA Family: Rosaceae A. f. var. obtusifolium, Ron A. f. var. fasciculatum., Riverside Co., A. Montalvo, RCRCD Vanderhoff (Creative Order: Rosales Commons CC) Subclass: Rosidae Class: Magnoliopsida A. Subspecific taxa 1. Adenostoma fasciculatum var. fasciculatum Hook. & Arn. 1. ADFAF 2. A. f. var. obusifolium S. Watson 2. ADFAO 3. A. f. var. prostratum Dunkle 3. (no NRCS code) B. Synonyms 1. A. f. var. densifolium Eastw. 2. A. brevifolium Nutt. 3. none. Formerly included as part of A. f. var. f. C. Common name 1. chamise, common chamise, California greasewood, greasewood, chamiso (Painter 2016) 2. San Diego chamise (Calflora 2016) 3. prostrate chamise (Calflora 2016) Phylogenetic studies using molecular sequence data placedAdenostoma closest to Chamaebatiaria and D. Taxonomic relationships Sorbaria (Morgan et al. 1994, Potter et al. 2007) and suggest tentative placement in subfamily Spiraeoideae, tribe Sorbarieae (Potter et al. 2007). E. Related taxa in region Adenostoma sparsifolium Torrey, known as ribbon-wood or red-shanks is the only other species of Adenostoma in California. It is a much taller, erect to spreading shrub of chaparral vegetation, often 2–6 m tall and has a more restricted distribution than A. fasciculatum. It occurs from San Luis Obispo Co. south into Baja California. Red-shanks produces longer, linear leaves on slender long shoots rather than having leaves clustered on short shoots (lacks "fascicled" leaves). Its bark is cinnamon-colored and in papery layers that sheds in long ribbons. F. Taxonomic issues The Jepson eFlora and the FNA recognize A. f. var. prostratum but the taxon is not recognized by USDA PLANTS (2016). -

Fort Ord Natural Reserve Plant List
UCSC Fort Ord Natural Reserve Plants Below is the most recently updated plant list for UCSC Fort Ord Natural Reserve. * non-native taxon ? presence in question Listed Species Information: CNPS Listed - as designated by the California Rare Plant Ranks (formerly known as CNPS Lists). More information at http://www.cnps.org/cnps/rareplants/ranking.php Cal IPC Listed - an inventory that categorizes exotic and invasive plants as High, Moderate, or Limited, reflecting the level of each species' negative ecological impact in California. More information at http://www.cal-ipc.org More information about Federal and State threatened and endangered species listings can be found at https://www.fws.gov/endangered/ (US) and http://www.dfg.ca.gov/wildlife/nongame/ t_e_spp/ (CA). FAMILY NAME SCIENTIFIC NAME COMMON NAME LISTED Ferns AZOLLACEAE - Mosquito Fern American water fern, mosquito fern, Family Azolla filiculoides ? Mosquito fern, Pacific mosquitofern DENNSTAEDTIACEAE - Bracken Hairy brackenfern, Western bracken Family Pteridium aquilinum var. pubescens fern DRYOPTERIDACEAE - Shield or California wood fern, Coastal wood wood fern family Dryopteris arguta fern, Shield fern Common horsetail rush, Common horsetail, field horsetail, Field EQUISETACEAE - Horsetail Family Equisetum arvense horsetail Equisetum telmateia ssp. braunii Giant horse tail, Giant horsetail Pentagramma triangularis ssp. PTERIDACEAE - Brake Family triangularis Gold back fern Gymnosperms CUPRESSACEAE - Cypress Family Hesperocyparis macrocarpa Monterey cypress CNPS - 1B.2, Cal IPC -

Oakmont Do Not Plant List
Oakmont Do Not Plant List Common Name Botanical Name Common Name Botanical Name Trees Shrubs/Vines Acacia Acacia spp. Bamboo ALL genera Arborvitae Thuja spp. Bluebeard Caryopteris spp. Australian tea tree Leptospermum Broom ALL genera laevigatum California buckwheat Eriogonum Black walnut Juglans nigra fasciculatum California bay Umbellularia Chamise or greasewood Adenostoma californica fasciculatum California pepper tree Schinus molle Chaparral pea Pickeringia montana Cedar Cedrus spp. Coyote brush Baccharis spp. Cypress Cupressus spp. Evergreen huckleberry Vaccinium ovatum Eucalyptus Eucalyptus spp. Gas plant Dictamus albus False cypress Chamaecyparis spp. Gorse Ulex europaeus Fir Abies spp. Honeysuckle Lonicera japonica ‘Halliana’ Giant chinquapin Chrysolepis chrysophylla Hopbush or hopseed Dodonaea viscosa bush Hemlock Tsuga spp. Juniper Juniperus spp. Honeylocust Gleditsia triacanthos Manzanita Arctostaphylos spp. Juniper Juniperus spp. (Ground cover variety okay) Leyland cypress Cupressus x leyandii (used as a hedge) New Zealand tea tree Leptospermum scoparium Palm ALL genera Rosemary Rosmarinus spp. Paperbark tree Melaleuca spp. Sagebrush Artemesia californica Pine Pinus spp. Scrub oak Quercus berberidifolia Spruce Picea spp. Grevillea Grevillea noellii Sweet gum Liquidambar Yew Taxus spp. (Also a styraciflua tree) Tamarisk Tamarix spp. Tan Oak Notholithocarpus densiflorus Tree of heaven Ailanthus altissima (Fourth Revision – 8/19/2018) (1/31/2021- Title and Spelling Corrections only) Common Name Botanical Name Grasses Fountain grass Pennisetum spp. Maiden grass Miscanthus sinensis Pampas grass Cortadaria selloana Ground Cover Big leaf periwinkle Vinca major Ivy Hedera spp. Juniper Juniperus spp. Mulch Woodchips and bark are not allowed in the 0–5- foot defensible space. The mulch types listed below are not allowed anywhere on residential property. Gorilla-hair Finely shredded redwood/cedar Rubber (Fourth Revision – 8/19/2018) (1/31/2021- Title and Spelling Corrections only) . -

A Trip to Study Oaks and Conifers in a Californian Landscape with the International Oak Society
A Trip to Study Oaks and Conifers in a Californian Landscape with the International Oak Society Harry Baldwin and Thomas Fry - 2018 Table of Contents Acknowledgments ....................................................................................................................................................... 3 Introduction .................................................................................................................................................................. 3 Aims and Objectives: .................................................................................................................................................. 4 How to achieve set objectives: ............................................................................................................................................. 4 Sharing knowledge of experience gained: ....................................................................................................................... 4 Map of Places Visited: ................................................................................................................................................. 5 Itinerary .......................................................................................................................................................................... 6 Background to Oaks .................................................................................................................................................... 8 Cosumnes River Preserve ........................................................................................................................................ -

Vascular Plants of the Coastal Dunes of Humboldt County, California
Humboldt State University Digital Commons @ Humboldt State University Botanical Studies Open Educational Resources and Data 4-2019 Vascular Plants of the Coastal Dunes of Humboldt County, California James P. Smith Jr Humboldt State University, [email protected] Follow this and additional works at: https://digitalcommons.humboldt.edu/botany_jps Part of the Botany Commons Recommended Citation Smith, James P. Jr, "Vascular Plants of the Coastal Dunes of Humboldt County, California" (2019). Botanical Studies. 41. https://digitalcommons.humboldt.edu/botany_jps/41 This Flora of Northwest California-Checklists of Local Sites is brought to you for free and open access by the Open Educational Resources and Data at Digital Commons @ Humboldt State University. It has been accepted for inclusion in Botanical Studies by an authorized administrator of Digital Commons @ Humboldt State University. For more information, please contact [email protected]. VASCULAR PLANTS OF THE COASTAL DUNES OF HUMBOLDT COUNTY, CALIFORNIA Compiled by James P. Smith, Jr. Professor Emeritus of Botany Department of Biological Sciences Humboldt State University Arcata, California Ninth Edition • August 2019 Amaryllidaceae — Onion or Amaryllis Family F E R N S Allium unifolium •One-leaved onion Dennstaedtiaceae — Bracken Fern Family Anacardiaceae — Cashew Family Pteridium aquilinum var. pubescens • Bracken fern Toxicodendron diversilobum • Poison-oak Ophioglossaceae — Adder’s-tongue Family Apocynaceae — Dogbane or Milkweed Family Botrychium multifidum • Leathery