(12) Patent Application Publication (10) Pub. No.: US 2006/0195944 A1 Heard Et Al
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Genome Annotation of Fritillaria Agrestis Bac Clone
GENOME ANNOTATION OF FRITILLARIA AGRESTIS BAC CLONE ___________________ A University Thesis Presented to the Faculty of California State University, East Bay ___________________ In Partial Fulfillment of the Requirements for the Degree Master of Science in Biological Science ___________________ By Rajhalutshimi Narayanaswamy September 2015 GENOME ANNOTATION OF FRITILLARJA AGRESTIS BAC CLONE By Rajhalutshimi Narayanaswamy Date: A~~u, ~o1s ii Acknowledgments I would like to thank my advisors Dr.Chris Baysdorfer, Dr.Claudia Udhe-Stone and Dr.Kenneth Curr for their support, guidance and encouragement that helped me to complete this project successfully. I would also like to thank my family members who have been supportive during the course of my project. iii Table of Contents Acknowledgments.............................................................................................................. iii List of Figures ................................................................................................................... vii 1 Introduction ...............................................................................................................1 1.1 Genome size variation ...........................................................................................1 1.2 Brief outline of the mechanisms of genome size variation ...................................2 1.3 Repetitious DNA ...................................................................................................3 1.4 Annotation ...........................................................................................................21 -

Botanical Priority Guidebook
Botanical Priority Protection Areas Alameda and Contra Costa Counties the East Bay Regional Park District. However, certain BPPAs include Hills have been from residential development. public parcels or properties with other conservation status. These are cases where land has been conserved since the creation of these boundaries or where potential management decisions have the poten- Following this initial mapping effort, the East Bay Chap- \ ntroduction tial to negatively affect an area’s botanical resources. Additionally, ter’s Conservation Committee began to utilize the con- each acre within these BPPAs represents a potential area of high pri- cept in draft form in key local planning efforts. Lech ority. Both urban and natural settings are included within these Naumovich, the chapter’s Conservation Analyst staff The lands that comprise the East Bay Chapter are located at the convergence boundaries, therefore, they are intended to be considered as areas person, showcased the map set in forums such as the of the San Francisco Bay, the North and South Coast Ranges, the Sacra- warranting further scrutiny due to the abundance of nearby sensitive BAOSC’s Upland Habitat Goals Project and the Green mento-San Joaquin Delta, and the San Joaquin Valley. The East Bay Chapter botanical resources supported by high quality habitat within each E A S T B A Y Vision Group (in association with Greenbelt Alliance); area supports a unique congregation of ecological conditions and native BPPA. Although a parcel, available for preservation through fee title C N P S East Bay Regional Park District’s Master Plan Process; plants. Based on historic botanical collections, the pressures from growth- purchase or conservation easement, may be located within the and local municipalities. -

Checklist of Vascular Plant Flora of Ventura County, California by David L
Checklist of Vascular Plant Flora of Ventura County, California By David L. Magney Abundance Scientific Name Common Name Habit Family Status Abies concolor (Gordon & Glendinning) Lindl. ex Hildebr. White Fir T Pinaceae U ? Abronia latifolia Eschsch. Coastal or Yellow Sand-verbena PH Nyctaginaceae X Abronia maritima Nutt. ex S. Watson Red or Sticky Sand-verbena, Beach PH Nyctaginaceae S, 4.2 Abronia maritima Nutt. ex S. Watson X A. umbellata Lam. Hybrid Sand-verbena AH Nyctaginaceae R Abronia neurophylla Standl. Beach Sand-verbena PH Nyctaginaceae R, T Abronia pogonantha Heimerl Desert Sand-verbena AH Nyctaginaceae R Abronia turbinata Torr. ex S. Watson Turbinate Sand-verbena A/PH Nyctaginaceae R Abronia umbellata Lam. ssp. umbellata Beach Sand-verbena PH Nyctaginaceae S Abronia villosa var. aurita (Abrams) Jeps. Woolly Sand-verbena AH Nyctaginaceae R, 1B.1 * Abutilon theophrasti Medikus Velvet Leaf AH Malvaceae R * Acacia baileyana F. Muell. Cootamundra Wattle S/T Fabaceae R * Acacia cultriforms A. Cunn. ex G. Don Sickle-leaved Acacia S Fabaceae R * Acacia dealbata Link Silver Wattle T Fabaceae R * Acacia longifolia (Andrews) Willd. Golden Wattle S/T Fabaceae R * Acacia retinodes Schldl. Everblooming Acacia T Fabaceae R * Acacia saligna (Labill.) H.L. Wendl. Golden Wreath Wattle S/T Fabaceae R Acamptopappus sphaerocephalus (Har. & Gray) Gray var. sphaerocephalus Rayless Goldenhead S Asteraceae R Acanthomintha obovata var. cordata Jokerst Heartleaf Thornmint AH Lamiaceae U, 1B.2 Acanthoscyphus parishii (Parry) Small var. parishii Parish Oxytheca AH Polygonaceae R, 4.2 Acanthoscyphus parishii var. abramsii (E.A. McGregor) Reveal Abrams Oxytheca AH Polygonaceae R, 1B.2 Acer macrophyllum Pursh Bigleaf Maple T Sapindaceae S Acer negundo var. -

Ukiah Western Hills Open Land Acquisition & Limited Development
Ukiah Western Hills Open Land Acquisition & Limited Development Agreement Draft Initial Study & Mitigated Negative Declaration Attachments April 16, 2021 ATTACHMENT A Existing Site Photographs Existing access road Existing water tank site Existing "house site" on one of the proposed Development Parcels ATTACHMENT B Prepared For: Michelle Irace, Planning Manager Department of Community Development 300 Seminary Avenue, Ukiah, CA 95482 APNs: 001-040-83, 157- 070-01, 157-070-02, and 003-190-01 Prepared by Jacobszoon & Associates, Inc. Alicia Ives Ringstad Senior Wildlife Biologist [email protected] Date: March 11, 2021 Updated: April 8, 2021 Biological Assessment Report Table of Contents Section 1.0: Introduction .................................................................................................................................................................. 2 Section 2.0: Regulations and Descriptions ....................................................................................................................................... 2 2.1 Regulatory Setting ................................................................................................................................................................. 2 2.2 Natural Communities and Sensitive Natural Communities .................................................................................................... 3 2.3 Special-Status Species........................................................................................................................................................... -

Table E-8. Bitter Creek NWR – Plants Bitter Creek NWR Scientific Name Common Name Family Acanthomintha Obovata Subsp
Appendix E - Plants and Wildlife Bitter Creek NWR Plant Lists Table E-8. Bitter Creek NWR – Plants Bitter Creek NWR Scientific Name Common Name Family Acanthomintha obovata subsp. cordata heart-leaved acanthomintha Lamiaceae Achillea millefolium common yarrow Asteraceae Acmispon americanus var. americanus [Lotus purshianus var. purshianus; Lotus typical American bird's-foot-trefoil Fabaceae unifoliolatus var. unifoliolatus] Acmispon brachycarpus [Lotus humistratus] short-podded lotus Fabaceae Acmispon glaber [Lotus scoparius] deer lotus Fabaceae Acmispon procumbens var. procumbens [Lotus typical silky bird's-foot-trefoil Fabaceae procumbens var. procumbens] Acmispon wrangelianus [Lotus wrangelianus, Wrangel's lotus Fabaceae Lotus subpinnatus, misapplied] Agoseris grandiflora var. grandiflora typical grassland agoseris Asteraceae Agoseris retrorsa spear-leaved agoseris Asteraceae Ailanthus altissima tree-of-heaven Simaroubaceae Aliciella leptomeria [Gilia leptomeria] sand aliciella Polemoniaceae Allium crispum crinkled onion Alliaceae [Liliaceae] Allium howellii var. howellii typical Howell's allium Alliaceae [Liliaceae] Allium peninsulare var. peninsulare typical peninsular allium Alliaceae [Liliaceae] Allophyllum gilioides subsp. gilioides typical gilia-like allophyllum Polemoniaceae Allophyllum gilioides subsp. violaceum gilia-like allophyllum Polemoniaceae Amaranthus blitoides mat amaranth Amaranthaceae Ambrosia acanthicarpa annual bur-sage Asteraceae Amsinckia douglasiana Douglas's fiddleneck Boraginaceae Amsinckia eastwoodiae Eastwood's -

Serpentine Endemism in the California Flora: a Database of Serpentine Affinity
MADRONÄ O, Vol. 52, No. 4, pp. 222±257, 2005 SERPENTINE ENDEMISM IN THE CALIFORNIA FLORA: A DATABASE OF SERPENTINE AFFINITY H. D. SAFFORD1,2,J.H.VIERS3, AND S. P. HARRISON2 1 USDA-Forest Service, Paci®c Southwest Region, 1323 Club Drive, Vallejo, CA 94592 [email protected] 2 Department of Environmental Science and Policy, University of California, Davis, CA 95616 3 Information Center for the Environment, DESP, University of California, Davis, CA 95616 ABSTRACT We present a summary of a database documenting levels of af®nity to ultrama®c (``serpentine'') sub- strates for taxa in the California ¯ora, USA. We constructed our database through an extensive literature search, expert opinion, ®eld observations, and intensive use of accession records at key herbaria. We developed a semi-quantitative methodology for determining levels of serpentine af®nity (strictly endemic, broadly endemic, strong ``indicator'', etc.) in the California ¯ora. In this contribution, we provide a list of taxa having high af®nity to ultrama®c/serpentine substrates in California, and present information on rarity, geographic distribution, taxonomy, and lifeform. Of species endemic to California, 12.5% are restricted to ultrama®c substrates. Most of these taxa come from a half-dozen plant families, and from only one or two genera within each family. The North Coast and Klamath Ranges support more serpentine endemics than the rest of the State combined. 15% of all plant taxa listed as threatened or endangered in California show some degree of association with ultrama®c substrates. Information in our database should prove valuable to efforts in ecology, ¯oristics, biosystematics, conservation, and land management. -

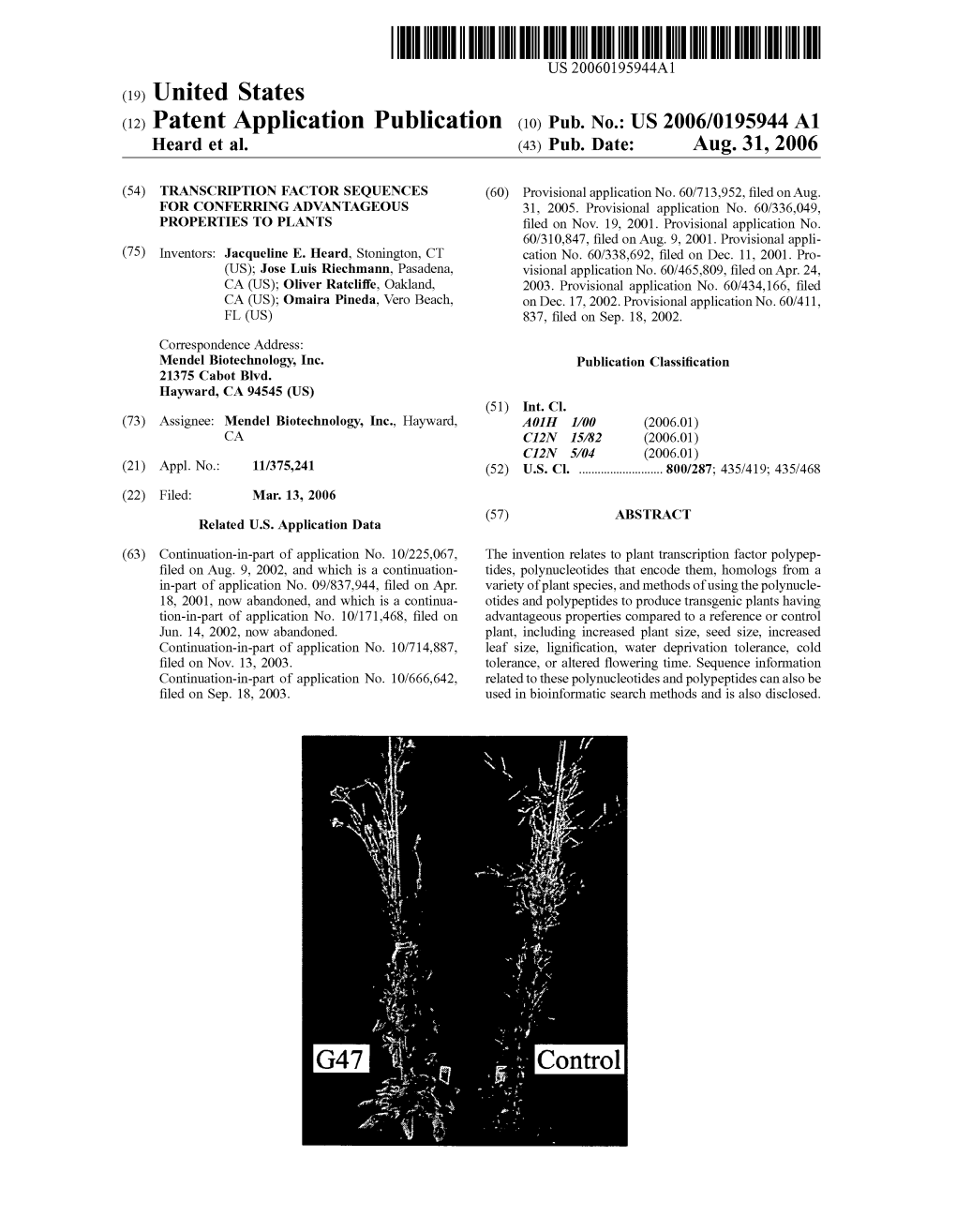
(12) United States Patent (10) Patent No.: US 7,598.429 B2 Heard Et Al
US007598429B2 (12) United States Patent (10) Patent No.: US 7,598.429 B2 Heard et al. (45) Date of Patent: *Oct. 6, 2009 (54) TRANSCRIPTION FACTOR SEQUENCES 6,329,567 B1 12/2001 Jofuku et al. FOR CONFERRING ADVANTAGEOUS 6,417,428 B1 7/2002 Thomashow et al. PROPERTIES TO PLANTS 6,664,446 B2 12/2003 Heard et al. 6,706,866 B1 3/2004 Thomashow et al. (75) Inventors: Jacqueline E. Heard, Stonington, CT 6,717,034 B2 4/2004 Jiang (US); Jose Luis Riechmann, Pasadena, 6.833,446 B1 12/2004 Wood et al. CA (US). Oliver Ratcliffe, Oakland, CA 6,835,540 B2 12/2004 Broun SS Omaira Pineda, Vero Beach, FL 6,846,669 B1 1/2005 Jofuku et al. (US) 6,946,586 B1 9, 2005 Fromm et al. (73) Assignee: Mendel Biotechnology, Inc., Hayward, 7,109,393 B2 9, 2006 Gutterson et al. CA (US) 7,135,616 B2 * 1 1/2006 Heard et al. ................ 800,278 7,196,245 B2 3/2007 Jiang et al. (*) Notice: Subject to any disclaimer, the term of this 7,223,904 B2 5/2007 Heard et al. patent is extended or adjusted under 35 7,238,860 B2 7/2007 Ratcliffe et al. U.S.C. 154(b) by 170 days. 7,345,217 B2 3/2008 Zhang et al. 2002fOO40490 A1 4/2002 Gorlach et al. This patent is subject to a terminal dis- 2003/0093837 A1 5.2003 Keddie et al. claimer. 2003/O121070 A1 6/2003 Adam et al. (21) Appl. No.: 11/375,241 2003. -

Studies in the Genus Fritillaria L. (Liliaceae)
Studies in the genus Fritillaria L. (Liliaceae) Peter D. Day, BSc. October 2017 Submitted in partial fulfilment of the requirements of the Degree of Doctor of Philosophy Supervisors: Prof. Andrew R. Leitch Dr Ilia J. Leitch Statement of Originality I, Peter Donal Day, confirm that the research included within this thesis is my own work or that where it has been carried out in collaboration with, or supported by others, that this is duly acknowledged below and my contribution indicated. Previously published material is also acknowledged below. I attest that I have exercised reasonable care to ensure that the work is original, and does not to the best of my knowledge break any UK law, infringe any third party’s copyright or other Intellectual Property Right, or contain any confidential material. I accept that the College has the right to use plagiarism detection software to check the electronic version of the thesis. I confirm that this thesis has not been previously submitted for the award of a degree by this or any other university. The copyright of this thesis rests with the author and no quotation from it or information derived from it may be published without the prior written consent of the author. Signature: Date: October 31st 2017 Details of collaboration and publications. Chapter 2 is published in Day et al. (2014): Day PD, Berger M, Hill L, Fay MF, Leitch AR, Leitch IJ, Kelly LJ (2014). Evolutionary relationships in the medicinally important genus Fritillaria L. (Liliaceae). Molecular Phylogenetics and Evolution, 80: 11-19. Martyn -

Development of Genomic Resources for Ornamental Lilies (Lilium L.)
Development of Genomic Resources for Ornamental Lilies (Lilium L.) Arwa Shahin Thesis committee Thesis supervisor Prof. dr. R.G.F. Visser Professor of Plant Breeding Wageningen University Thesis co-supervisors Dr. ir. J. M. van Tuyl Senior Scientist, Plant Research International Wageningen University and Research Centre Dr. P.F.P. Arens Scientist, Plant Research International Wageningen University and Research Centre Other members Prof. dr. ir. H.J. Bouwmeester, Wageningen University Prof. dr. E.J. Woltering, Wageningen University Dr. ir. S. Heimovaara, Royal van Zanten, Rijsenhout, the Netherlands Dr. ir. S.A. Peters, Plant Research International, Wageningen This research was conducted under the auspices of the Graduate School of Experimental Plant Science. II Development of Genomic Resources for Ornamental Lilies (Lilium L.) Arwa Shahin Thesis submitted in fulfillment of the requirements for the degree of doctor at Wageningen University by the authority of the Rector Magnificus Prof. dr. M.J. Kropff, in the presence of the Thesis Committee appointed by the Academic Board to be defended in public on Tuesday 19th June, 2012 at 4 p.m. in the Aula. III Shahin A. Development of Genomic Resources for Ornamental Lilies (Lilium L.). 169 pages. Thesis, Wageningen University, Wageningen, NL (2012) With references, with summaries in Dutch and English ISBN 978-94-6173-300-9 IV Table of contents Chapter 1 General introduction 1 - 13 Chapter 2 Genetic mapping in Lilium L.: mapping of major genes and 15-32 QTLs for several ornamental traits and disease resistances Chapter 3 SNP markers retrieval for a non-model species: A practical 33-49 approach Chapter 4 Generation and analysis of expressed sequence tags in the 51-68 extreme large genomes Lilium L. -

An Updated Database of Serpentine Endemism in the California Flora
AN UPDATED DATABASE OF SERPENTINE ENDEMISM IN THE CALIFORNIA FLORA Authors: Safford, Hugh, and Miller, Jesse E. D. Source: Madroño, 67(2) : 85-104 Published By: California Botanical Society URL: https://doi.org/10.3120/0024-9637-67.2.85 BioOne Complete (complete.BioOne.org) is a full-text database of 200 subscribed and open-access titles in the biological, ecological, and environmental sciences published by nonprofit societies, associations, museums, institutions, and presses. Your use of this PDF, the BioOne Complete website, and all posted and associated content indicates your acceptance of BioOne’s Terms of Use, available at www.bioone.org/terms-of-use. Usage of BioOne Complete content is strictly limited to personal, educational, and non - commercial use. Commercial inquiries or rights and permissions requests should be directed to the individual publisher as copyright holder. BioOne sees sustainable scholarly publishing as an inherently collaborative enterprise connecting authors, nonprofit publishers, academic institutions, research libraries, and research funders in the common goal of maximizing access to critical research. Downloaded From: https://bioone.org/journals/Madroño on 16 Oct 2020 Terms of Use: https://bioone.org/terms-of-use Access provided by California Botanical Society MADRONO˜ , Vol. 67, No. 2, pp. 85–104, 2020 AN UPDATED DATABASE OF SERPENTINE ENDEMISM IN THE CALIFORNIA FLORA HUGH SAFFORD USDA Forest Service, Pacific Southwest Region, 1323 Club Drive, Vallejo, CA 94592; Department of Environmental Science and Policy, University of California, Davis, CA 95616 [email protected] JESSE E. D. MILLER Department of Biology, Stanford University, Palo Alto, CA 94305 ABSTRACT We update and revise the serpentine affinity database of Safford et al. -

VOL 20 NO4 Oct-Dec 2013
VOL 20 NO 4 Oct - Dec 2013 ISSN 1727-2874 News & Short Communications Consultancy Services – A New and Little-Known Role of a Pharmacist An Overview of Contemporary Treatment for Rheumatoid Arthritis (2 CE Units) Over-the-Counter Options for Diarrhea in Children Distinction of the Bulbs of Fritillariae thunbergii Miq., Fritillariae ussuriensis Maxim and Fritillariae huperhensis Hsiao et K.C. Hsia by Concert Techniques Concert Approach to the Authentication, Qualitative Evaluation and Bioactivity Assessment of Beimu Invitation letter Hong Kong Pharmacy Conference 2014 The Annual General Meeting of the Pharmaceutical Society of Hong Kong and the 20th Anniversary of the Hong Kong Pharmaceutical Journal A weekend trip to Taipei with the President of PSHK ACTILYSE®/ENANTONE® 6 MONTH DPS 30MG Editorial An Opportunity to Turn the Tide – My Ponder After Eighteen Years of Association with HKPJ I am very pleased to present to will always be there. Fortunately, starting from last summer you the latest issue of Hong Kong we have and will continue to have more qualifi ed pharmacists Pharmaceutical Journal (HKPJ), graduating from both local pharmacy schools than ever before. which has a history of more than This means that the pharmacy community will have a steady twenty years by this month. From growth in term of number from now on. This phenomenon may the viewpoint of growth of a human gradually solve the problem as more number of registered being, it is regarded reaching a stage pharmacists making their endeavor. When both pharmacy of mature. I believe no one would schools grow and stabilized, academic staff in these two doubt that HKPJ is the only local schools should also spare some efforts and manpower to keep pharmaceutical journal as there is no this journal running. -

2Nd Edition) California Native Plant Society April 1980 COUNTY and ISLAND CODES
INVENTORY of RARE AND ENDANGERED VASCULAR PLANTS of CALIFORNIA , Special Publication No. 1 (2nd Edition) California Native Plant Society April 1980 COUNTY AND ISLAND CODES 1 Alameda 35 San Benito 2 Alpine 36 San Bernardino 3 Amador 37 San Diego 4 Butte 38 San Francisco 5 Calaveras 39 San Joaquin 6 Colusa 40 San Luis Obispo 7 Contra Costa 41 San Mateo 8 Del orte 42 Santa Barbara 9 El Dorado 43 Santa Clara 10 Fresno 44 Santa Cruz 11 Glenn 45 Shasta 12 Humboldt 46 Sierra 13 Imperial 47 Siskiyou 14 Inyo 48 Solano 15 Kern 49 Sonoma 16 Kings 50 Stanislaus 17 Lake 51 Sutter 18 Lassen 52 Tehama 19 Los Angeles 53 Trinity 20 Madera 54 Tulare 21 Marin 55 Tuolumne 22 Mariposa 56 Ventura 23 Mendocino 57 Yolo 24 Merced 58 Yuba 25 Modoc 59 Anacapa Islands (Ventura County) 26 Mono 60 San Clemente Island (Los Angeles County) 27 Monterey 61 San Miguel Island (Santa Barbara County) 28 Napa 62 San Nicolas Island (Ventura County) 29 Nevada 63 Santa Barbara Island (Santa Barbara County) 30 Orange 64 Santa Catalina Island (Los Angeles County) 31 Placer 65 Santa Cruz Island (Santa Barbara County) 32 Plumas 66 Santa Rosa Island (Santa Barbara County) 33 Rivers;de 67 Farallon Islands (San Francisco County) 34 Sacramento ABBREVIATIONS AND SYMBOLS AZ -Arizona SO-Sonora, Mexico BA -Baja California, Mexico ST-Smithsonian threatened plant CE -California endangered plant SX-Smithsonian extinct plant CR -California rare plant FL -federally listed plant GU-Isla Guadalupe, Baja California NV - evada +-this state and beyond OR-Oregon ++-widespread outside California SE -Smithsonian endangered plant *-extinct or extirpated The cover illustration of Rai//ardella pringlei, a Trinity and Siskiyou Co.