Eya1-Dependent Homeostasis of Morphogenetic Territories During Collective Cell Migration
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Identification of Candidate Genes and Pathways Associated with Obesity
animals Article Identification of Candidate Genes and Pathways Associated with Obesity-Related Traits in Canines via Gene-Set Enrichment and Pathway-Based GWAS Analysis Sunirmal Sheet y, Srikanth Krishnamoorthy y , Jihye Cha, Soyoung Choi and Bong-Hwan Choi * Animal Genome & Bioinformatics, National Institute of Animal Science, RDA, Wanju 55365, Korea; [email protected] (S.S.); [email protected] (S.K.); [email protected] (J.C.); [email protected] (S.C.) * Correspondence: [email protected]; Tel.: +82-10-8143-5164 These authors contributed equally. y Received: 10 October 2020; Accepted: 6 November 2020; Published: 9 November 2020 Simple Summary: Obesity is a serious health issue and is increasing at an alarming rate in several dog breeds, but there is limited information on the genetic mechanism underlying it. Moreover, there have been very few reports on genetic markers associated with canine obesity. These studies were limited to the use of a single breed in the association study. In this study, we have performed a GWAS and supplemented it with gene-set enrichment and pathway-based analyses to identify causative loci and genes associated with canine obesity in 18 different dog breeds. From the GWAS, the significant markers associated with obesity-related traits including body weight (CACNA1B, C22orf39, U6, MYH14, PTPN2, SEH1L) and blood sugar (PRSS55, GRIK2), were identified. Furthermore, the gene-set enrichment and pathway-based analysis (GESA) highlighted five enriched pathways (Wnt signaling pathway, adherens junction, pathways in cancer, axon guidance, and insulin secretion) and seven GO terms (fat cell differentiation, calcium ion binding, cytoplasm, nucleus, phospholipid transport, central nervous system development, and cell surface) which were found to be shared among all the traits. -

Mirc11 Disrupts Inflammatory but Not Cytotoxic Responses of NK Cells
Published OnlineFirst September 12, 2019; DOI: 10.1158/2326-6066.CIR-18-0934 Research Article Cancer Immunology Research Mirc11 Disrupts Inflammatory but Not Cytotoxic Responses of NK Cells Arash Nanbakhsh1, Anupallavi Srinivasamani1, Sandra Holzhauer2, Matthew J. Riese2,3,4, Yongwei Zheng5, Demin Wang4,5, Robert Burns6, Michael H. Reimer7,8, Sridhar Rao7,8, Angela Lemke9,10, Shirng-Wern Tsaih9,10, Michael J. Flister9,10, Shunhua Lao1,11, Richard Dahl12, Monica S. Thakar1,11, and Subramaniam Malarkannan1,3,4,9,11 Abstract Natural killer (NK) cells generate proinflammatory cyto- g–dependent clearance of Listeria monocytogenes or B16F10 kines that are required to contain infections and tumor melanoma in vivo by NK cells. These functional changes growth. However, the posttranscriptional mechanisms that resulted from Mirc11 silencing ubiquitin modifiers A20, regulate NK cell functions are not fully understood. Here, we Cbl-b, and Itch, allowing TRAF6-dependent activation of define the role of the microRNA cluster known as Mirc11 NF-kB and AP-1. Lack of Mirc11 caused increased translation (which includes miRNA-23a, miRNA-24a, and miRNA-27a) of A20, Cbl-b, and Itch proteins, resulting in deubiquityla- in NK cell–mediated proinflammatory responses. Absence tion of scaffolding K63 and addition of degradative K48 of Mirc11 did not alter the development or the antitumor moieties on TRAF6. Collectively, our results describe a func- cytotoxicity of NK cells. However, loss of Mirc11 reduced tion of Mirc11 that regulates generation of proinflammatory generation of proinflammatory factors in vitro and interferon- cytokines from effector lymphocytes. Introduction TRAF2 and TRAF6 promote K63-linked polyubiquitination that is required for subcellular localization of the substrates (20), Natural killer (NK) cells generate proinflammatory factors and and subsequent activation of NF-kB (21) and AP-1 (22). -

Genetic Alterations of Protein Tyrosine Phosphatases in Human Cancers
Oncogene (2015) 34, 3885–3894 © 2015 Macmillan Publishers Limited All rights reserved 0950-9232/15 www.nature.com/onc REVIEW Genetic alterations of protein tyrosine phosphatases in human cancers S Zhao1,2,3, D Sedwick3,4 and Z Wang2,3 Protein tyrosine phosphatases (PTPs) are enzymes that remove phosphate from tyrosine residues in proteins. Recent whole-exome sequencing of human cancer genomes reveals that many PTPs are frequently mutated in a variety of cancers. Among these mutated PTPs, PTP receptor T (PTPRT) appears to be the most frequently mutated PTP in human cancers. Beside PTPN11, which functions as an oncogene in leukemia, genetic and functional studies indicate that most of mutant PTPs are tumor suppressor genes. Identification of the substrates and corresponding kinases of the mutant PTPs may provide novel therapeutic targets for cancers harboring these mutant PTPs. Oncogene (2015) 34, 3885–3894; doi:10.1038/onc.2014.326; published online 29 September 2014 INTRODUCTION tyrosine/threonine-specific phosphatases. (4) Class IV PTPs include Protein tyrosine phosphorylation has a critical role in virtually all four Drosophila Eya homologs (Eya1, Eya2, Eya3 and Eya4), which human cellular processes that are involved in oncogenesis.1 can dephosphorylate both tyrosine and serine residues. Protein tyrosine phosphorylation is coordinately regulated by protein tyrosine kinases (PTKs) and protein tyrosine phosphatases 1 THE THREE-DIMENSIONAL STRUCTURE AND CATALYTIC (PTPs). Although PTKs add phosphate to tyrosine residues in MECHANISM OF PTPS proteins, PTPs remove it. Many PTKs are well-documented oncogenes.1 Recent cancer genomic studies provided compelling The three-dimensional structures of the catalytic domains of evidence that many PTPs function as tumor suppressor genes, classical PTPs (RPTPs and non-RPTPs) are extremely well because a majority of PTP mutations that have been identified in conserved.5 Even the catalytic domain structures of the dual- human cancers are loss-of-function mutations. -

Phosphatome Rnai Screen Identifies Eya1 As a Positive Regulator of Hedgehog Signal Transduction
Phosphatome RNAi Screen Identifies Eya1 as a Positive Regulator of Hedgehog Signal Transduction The Harvard community has made this article openly available. Please share how this access benefits you. Your story matters Citation Eisner, Adriana. 2013. Phosphatome RNAi Screen Identifies Eya1 as a Positive Regulator of Hedgehog Signal Transduction. Doctoral dissertation, Harvard University. Citable link http://nrs.harvard.edu/urn-3:HUL.InstRepos:11181213 Terms of Use This article was downloaded from Harvard University’s DASH repository, and is made available under the terms and conditions applicable to Other Posted Material, as set forth at http:// nrs.harvard.edu/urn-3:HUL.InstRepos:dash.current.terms-of- use#LAA Phosphatome RNAi Screen Identifies Eya1 as a Positive Regulator of Hedgehog Signal Transduction A dissertation presented by Adriana Eisner to The Division of Medical Sciences in partial fulfillment of the requirements for the degree of Doctor of Philosophy in the subject of Neurobiology Harvard University Cambridge, Massachusetts June 2013 © 2013 Adriana Eisner All rights reserved. Dissertation Advisor: Dr. Rosalind Segal Adriana Eisner Phosphatome RNAi Screen Identifies Eya1 as a Positive Regulator of Hedgehog Signal Transduction Abstract The Hedgehog (Hh) signaling pathway is vital for vertebrate embryogenesis and aberrant activation of the pathway can cause tumorigenesis in humans. In this study, we used a phosphatome RNAi screen for regulators of Hh signaling to identify a member of the Eyes Absent protein family, Eya1, as a positive regulator of Hh signal transduction. Eya1 is both a phosphatase and transcriptional regulator. Eya family members have been implicated in tumor biology, and Eya1 is highly expressed in a particular subtype of medulloblastoma (MB). -

Genetic Testing Medical Policy – Genetics
Genetic Testing Medical Policy – Genetics Please complete all appropriate questions fully. Suggested medical record documentation: • Current History & Physical • Progress Notes • Family Genetic History • Genetic Counseling Evaluation *Failure to include suggested medical record documentation may result in delay or possible denial of request. PATIENT INFORMATION Name: Member ID: Group ID: PROCEDURE INFORMATION Genetic Counseling performed: c Yes c No **Please check the requested analyte(s), identify number of units requested, and provide indication/rationale for testing. 81400 Molecular Pathology Level 1 Units _____ c ACADM (acyl-CoA dehydrogenase, C-4 to C-12 straight chain, MCAD) (e.g., medium chain acyl dehydrogenase deficiency), K304E variant _____ c ACE (angiotensin converting enzyme) (e.g., hereditary blood pressure regulation), insertion/deletion variant _____ c AGTR1 (angiotensin II receptor, type 1) (e.g., essential hypertension), 1166A>C variant _____ c BCKDHA (branched chain keto acid dehydrogenase E1, alpha polypeptide) (e.g., maple syrup urine disease, type 1A), Y438N variant _____ c CCR5 (chemokine C-C motif receptor 5) (e.g., HIV resistance), 32-bp deletion mutation/794 825del32 deletion _____ c CLRN1 (clarin 1) (e.g., Usher syndrome, type 3), N48K variant _____ c DPYD (dihydropyrimidine dehydrogenase) (e.g., 5-fluorouracil/5-FU and capecitabine drug metabolism), IVS14+1G>A variant _____ c F13B (coagulation factor XIII, B polypeptide) (e.g., hereditary hypercoagulability), V34L variant _____ c F2 (coagulation factor 2) (e.g., -

Open Chromatin Dynamics in Prosensory Cells of the Embryonic Mouse Cochlea Received: 10 February 2019 Brent A
www.nature.com/scientificreports OPEN Open chromatin dynamics in prosensory cells of the embryonic mouse cochlea Received: 10 February 2019 Brent A. Wilkerson1,2, Alex D. Chitsazan1,2,3, Leah S. VandenBosch1,4, Matthew S. Wilken1,5, Accepted: 10 June 2019 Thomas A. Reh1,2 & Olivia Bermingham-McDonogh1,2 Published: xx xx xxxx Hearing loss is often due to the absence or the degeneration of hair cells in the cochlea. Understanding the mechanisms regulating the generation of hair cells may therefore lead to better treatments for hearing disorders. To elucidate the transcriptional control mechanisms specifying the progenitor cells (i.e. prosensory cells) that generate the hair cells and support cells critical for hearing function, we compared chromatin accessibility using ATAC-seq in sorted prosensory cells (Sox2-EGFP+) and surrounding cells (Sox2-EGFP−) from E12, E14.5 and E16 cochlear ducts. In Sox2-EGFP+, we fnd greater accessibility in and near genes restricted in expression to the prosensory region of the cochlear duct including Sox2, Isl1, Eya1 and Pou4f3. Furthermore, we fnd signifcant enrichment for the consensus binding sites of Sox2, Six1 and Gata3—transcription factors required for prosensory development—in the open chromatin regions. Over 2,200 regions displayed diferential accessibility with developmental time in Sox2-EGFP+ cells, with most changes in the E12-14.5 window. Open chromatin regions detected in Sox2-EGFP+ cells map to over 48,000 orthologous regions in the human genome that include regions in genes linked to deafness. Our results reveal a dynamic landscape of open chromatin in prosensory cells with potential implications for cochlear development and disease. -

Phosphatases Page 1
Phosphatases esiRNA ID Gene Name Gene Description Ensembl ID HU-05948-1 ACP1 acid phosphatase 1, soluble ENSG00000143727 HU-01870-1 ACP2 acid phosphatase 2, lysosomal ENSG00000134575 HU-05292-1 ACP5 acid phosphatase 5, tartrate resistant ENSG00000102575 HU-02655-1 ACP6 acid phosphatase 6, lysophosphatidic ENSG00000162836 HU-13465-1 ACPL2 acid phosphatase-like 2 ENSG00000155893 HU-06716-1 ACPP acid phosphatase, prostate ENSG00000014257 HU-15218-1 ACPT acid phosphatase, testicular ENSG00000142513 HU-09496-1 ACYP1 acylphosphatase 1, erythrocyte (common) type ENSG00000119640 HU-04746-1 ALPL alkaline phosphatase, liver ENSG00000162551 HU-14729-1 ALPP alkaline phosphatase, placental ENSG00000163283 HU-14729-1 ALPP alkaline phosphatase, placental ENSG00000163283 HU-14729-1 ALPPL2 alkaline phosphatase, placental-like 2 ENSG00000163286 HU-07767-1 BPGM 2,3-bisphosphoglycerate mutase ENSG00000172331 HU-06476-1 BPNT1 3'(2'), 5'-bisphosphate nucleotidase 1 ENSG00000162813 HU-09086-1 CANT1 calcium activated nucleotidase 1 ENSG00000171302 HU-03115-1 CCDC155 coiled-coil domain containing 155 ENSG00000161609 HU-09022-1 CDC14A CDC14 cell division cycle 14 homolog A (S. cerevisiae) ENSG00000079335 HU-11533-1 CDC14B CDC14 cell division cycle 14 homolog B (S. cerevisiae) ENSG00000081377 HU-06323-1 CDC25A cell division cycle 25 homolog A (S. pombe) ENSG00000164045 HU-07288-1 CDC25B cell division cycle 25 homolog B (S. pombe) ENSG00000101224 HU-06033-1 CDKN3 cyclin-dependent kinase inhibitor 3 ENSG00000100526 HU-02274-1 CTDSP1 CTD (carboxy-terminal domain, -

The Evolving (Epi)Genetic Landscape of Pancreatic Neuroendocrine Tumours
26 9 Endocrine-Related C P Pipinikas et al. Evolution of PanNET genomics 26:9 R519–R544 Cancer REVIEW The evolving (epi)genetic landscape of pancreatic neuroendocrine tumours Christodoulos P Pipinikas1, Alison M Berner1, Teresa Sposito1 and Christina Thirlwell1,2 1Medical Genomics Laboratory, University College London Cancer Institute, London, UK 2Neuroendocrine Tumour Unit and Department of Oncology, Royal Free Hospital, London, UK Correspondence should be addressed to C Thirlwell: [email protected] Abstract Neuroendocrine neoplasms (NENs) are a relatively rare group of heterogeneous tumours Key Words originating from neuroendocrine cells found throughout the body. Pancreatic NENs f pancreatic neuroendocrine (PanNENs) are the second most common pancreatic malignancy accounting for 1–3% of tumour all neoplasms developing in the pancreas. Despite having a low background mutation f MEN1 rate, driver mutations in MEN1, DAXX/ATRX and mTOR pathway genes (PTEN, TSC1/2) are f DAXX implicated in disease development and progression. Their increased incidence coupled f ATRX with advances in sequencing technologies has reignited the interest in PanNEN research f mTOR and has accelerated the acquisition of molecular data. Studies utilising such technological f MUTYH advances have further enriched our knowledge of PanNENs’ biology through novel f BRCA2 findings, including higher-than-expected presence of germline mutations in 17% of f CHEK2 sporadic tumours of no familial background, identification of novel mutational signatures f alternative lengthening of telomeres and complex chromosomal rearrangements and a dysregulated epigenetic machinery. f chromatin remodelling. Integrated genomic studies have progressed the field by identifying the synergistic action between different molecular mechanisms, while holding the promise for deciphering disease heterogeneity. -

Development in the Mammalian Auditory System Depends on Transcription Factors
International Journal of Molecular Sciences Review Development in the Mammalian Auditory System Depends on Transcription Factors Karen L. Elliott 1, Gabriela Pavlínková 2 , Victor V. Chizhikov 3, Ebenezer N. Yamoah 4 and Bernd Fritzsch 1,* 1 Department of Biology, University of Iowa, Iowa City, IA 52242, USA; [email protected] 2 Institute of Biotechnology of the Czech Academy of Sciences, 25250 Vestec, Czechia; [email protected] 3 Department of Anatomy and Neurobiology, The University of Tennessee Health Science Center, Memphis, TN 38163, USA; [email protected] 4 Department of Physiology and Cell Biology, School of Medicine, University of Nevada, Reno, NV 89557, USA; [email protected] * Correspondence: [email protected] Abstract: We review the molecular basis of several transcription factors (Eya1, Sox2), including the three related genes coding basic helix–loop–helix (bHLH; see abbreviations) proteins (Neurog1, Neurod1, Atoh1) during the development of spiral ganglia, cochlear nuclei, and cochlear hair cells. Neuronal development requires Neurog1, followed by its downstream target Neurod1, to cross- regulate Atoh1 expression. In contrast, hair cells and cochlear nuclei critically depend on Atoh1 and require Neurod1 expression for interactions with Atoh1. Upregulation of Atoh1 following Neurod1 loss changes some vestibular neurons’ fate into “hair cells”, highlighting the significant interplay between the bHLH genes. Further work showed that replacing Atoh1 by Neurog1 rescues some hair cells from complete absence observed in Atoh1 null mutants, suggesting that bHLH genes can partially replace Citation: Elliott, K.L.; Pavlínková, G.; one another. The inhibition of Atoh1 by Neurod1 is essential for proper neuronal cell fate, and in Chizhikov, V.V.; Yamoah, E.N.; the absence of Neurod1, Atoh1 is upregulated, resulting in the formation of “intraganglionic” HCs. -

Lineage-Specific Effector Signatures of Invariant NKT Cells Are Shared Amongst Δγ T, Innate Lymphoid, and Th Cells
Downloaded from http://www.jimmunol.org/ by guest on September 26, 2021 δγ is online at: average * The Journal of Immunology , 10 of which you can access for free at: 2016; 197:1460-1470; Prepublished online 6 July from submission to initial decision 4 weeks from acceptance to publication 2016; doi: 10.4049/jimmunol.1600643 http://www.jimmunol.org/content/197/4/1460 Lineage-Specific Effector Signatures of Invariant NKT Cells Are Shared amongst T, Innate Lymphoid, and Th Cells You Jeong Lee, Gabriel J. Starrett, Seungeun Thera Lee, Rendong Yang, Christine M. Henzler, Stephen C. Jameson and Kristin A. Hogquist J Immunol cites 41 articles Submit online. Every submission reviewed by practicing scientists ? is published twice each month by Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts http://jimmunol.org/subscription http://www.jimmunol.org/content/suppl/2016/07/06/jimmunol.160064 3.DCSupplemental This article http://www.jimmunol.org/content/197/4/1460.full#ref-list-1 Information about subscribing to The JI No Triage! Fast Publication! Rapid Reviews! 30 days* Why • • • Material References Permissions Email Alerts Subscription Supplementary The Journal of Immunology The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 2016 by The American Association of Immunologists, Inc. All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. This information is current as of September 26, 2021. The Journal of Immunology Lineage-Specific Effector Signatures of Invariant NKT Cells Are Shared amongst gd T, Innate Lymphoid, and Th Cells You Jeong Lee,* Gabriel J. -
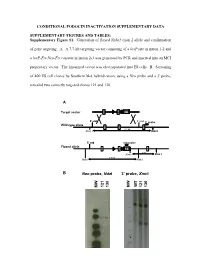
Supplementary Figure S1. Generation of Floxed Nphs2 Exon 2 Allele and Confirmation
CONDITIONAL PODOCIN INACTIVATION SUPPLEMENTARY DATA SUPPLEMENTARY FIGURES AND TABLES: Supplementary Figure S1. Generation of floxed Nphs2 exon 2 allele and confirmation of gene targeting. A. A 7.7-kb targeting vector consisting of a loxP site in intron 1-2 and a loxP-Frt-Neo-Frt cassette in intron 2-3 was generated by PCR and inserted into an MCI proprietary vector. The linearized vector was electroporated into ES cells. B. Screening of 400 ES cell clones by Southern blot hybridization, using a Neo probe and a 3' probe, revealed two correctly targeted clones 121 and 130. A Target vector 1 2 NEO 5’ end 3’ end 3’ probe Wild-type allele 1 2 3 9.2 kb Xmn I Xmn I 5’ end neo probe Floxed allele 1 2 NEO 3 5 kb Xmn I Xmn I 7.8 kb Nde I Nde I B Neo probe, NdeI 3’ probe, XmnI WT 121 130 121 130 MW MW - 1 - Supplementary Figure S2. Generation of Nphs2lox2/-,Cre+ mice and excision of exon 2 upon Cre recombinase induction. A. Triallelic Nphs2lox2/-,Cre+ mice were obtained by mating phenotypically normal Nphs2lox2/lox2 mice with Nphs2+/-,Cre+ mice. Mendelian inheritance of these alleles was observed. B. Genotypes were verified by multiplex PCR of tail genomic DNA. C. Cre recombinase activity was induced upon tamoxifen administration, leading to excision of the floxed exon 2 of the Nphs2 gene. D. Cre recombinase activity in the kidney was verified by PCR using a set of forward and reverse primers designed around exon 2 and demonstrating a 692-bp product (before Cre) and a 316-bp product (after Cre) using genomic DNA extracted from the renal cortex. -
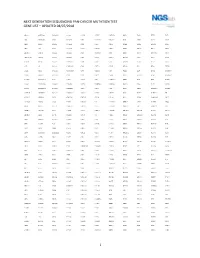
1 Next Generation Sequencing Pan-Cancer Mutation Test Gene List – Updated 08/07/2018
NEXT GENERATION SEQUENCING PAN-CANCER MUTATION TEST GENE LIST – UPDATED 08/07/2018 ABCC3 ANKRD26 BAIAP2L1 C11orf1 CCT6B CENPU CREB3L2 DDX6 EGR2 ETV5 FGF3 ABI1 ANKRD28 BAP1 C11orf30 CD19 CEP170B CREBBP DEK EGR3 ETV6 FGF4 ABL1 ANLN BARD1 C11orf54 CD22 CEP57 CRKL DGKB EGR4 EWSR1 FGF5 ABL2 APC BAX C11orf95 CD274 CEP85L CRLF2 DGKI EIF4A2 EXO1 FGF6 ABLIM1 APH1A BAZ2A C2CD2L CD28 CHCHD7 CRTC1 DGKZ EIF4E EXOSC6 FGF7 ABRAXAS1 APLP2 BCAS3 C2orf44 CD36 CHD2 CRTC3 DICER1 ELF4 EXT1 FGF8 ACACA APOD BCAS4 CACNA1F CD44 CHD6 CSF1 DIRAS3 ELK4 EXT2 FGF9 ACE AR BCL10 CACNA1G CD58 CHEK1 CSF1R DIS3L2 ELL EYA1 FGFR1 ACER1 ARAF BCL11A CACNA2D3 CD70 CHEK2 CSF3 DKK1 ELN EYA2 FGFR1OP ACKR3 ARFRP1 BCL11B CAD CD74 CHIC2 CSF3R DKK2 ELOVL2 EZH2 FGFR1OP2 ACSBG1 ARHGAP20 BCL2 CALR CD79A CHL1 CSNK1G2 DKK4 ELP2 EZR FGFR2 ACSL3 ARHGAP26 BCL2A1 CAMK2A CD79B CHMP2B CSNK2A1 DLEC1 EML1 FAF1 FGFR3 ACSL6 ARHGEF12 BCL2L1 CAMK2B CD8A CHN1 CTCF DLL1 EML4 FAM127C FGFR4 ACVR1B ARHGEF7 BCL2L2 CAMK2G CDC14A CHST11 CTDSP2 DLL3 ENPP2 FAM19A2 FH ACVR1C ARID1A BCL3 CAMTA1 CDC14B CHUK CTLA4 DLL4 EP300 FAM19A5 FHIT ACVR2A ARID2 BCL6 CANT1 CDC25A CIC CTNNA1 DMRT1 EP400 FAM46C FHL2 ADD3 ARIH2 BCL7A CAPRIN1 CDC25C CIITA CTNNB1 DMRTA2 EPC1 FAM64A FIGF ADGRA2 ARL6IP5 BCL9 CAPZB CDC42 CIRH1A CTNND2 DNAJB1 EPCAM FANCA FIP1L1 ADGRG7 ARNT BCOR CARD11 CDC73 CIT CTRB1 DNM1 EPHA10 FANCB FLCN ADM ARRDC4 BCORL1 CARM1 CDH1 CKB CTSA DNM2 EPHA2 FANCC FLI1 AFF1 ASMTL BCR CARS CDH11 CKS1B CUL4A DNM3 EPHA3 FANCD2 FLNA AFF3 ASPH BDNF CASC5 CDK1 CLP1 CUL4B DNMT1 EPHA5 FANCE FLNC AFF4 ASPSCR1 BHLHE22 CASP3 CDK12