Plasmid Replication-Associated Single-Strand-Specific
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Replisome Assembly at Oric, the Replication Origin of E. Coli, Reveals an Explanation for Initiation Sites Outside an Origin
Molecular Cell, Vol. 4, 541±553, October, 1999, Copyright 1999 by Cell Press Replisome Assembly at oriC, the Replication Origin of E. coli, Reveals an Explanation for Initiation Sites outside an Origin Linhua Fang,*§ Megan J. Davey,² and Mike O'Donnell²³ have not been addressed. For example, is the local un- *Microbiology Department winding sufficiently large for two helicases to assemble Joan and Sanford I. Weill Graduate School of Medical for bidirectional replication, or does one helicase need Sciences of Cornell University to enter first and expand the bubble via helicase action New York, New York 10021 to make room for the second helicase? The known rep- ² The Rockefeller University and licative helicases are hexameric and encircle ssDNA. Howard Hughes Medical Institute Which strand does the initial helicase(s) at the origin New York, New York 10021 encircle, and if there are two, how are they positioned relative to one another? Primases generally require at least transient interaction with helicase to function. Can Summary primase function with the helicase(s) directly after heli- case assembly at the origin, or must helicase-catalyzed This study outlines the events downstream of origin DNA unwinding occur prior to RNA primer synthesis? unwinding by DnaA, leading to assembly of two repli- Chromosomal replicases are comprised of a ring-shaped cation forks at the E. coli origin, oriC. We show that protein clamp that encircles DNA, a clamp-loading com- two hexamers of DnaB assemble onto the opposing plex that uses ATP to assemble the clamp around DNA, strands of the resulting bubble, expanding it further, and a DNA polymerase that binds the circular clamp, yet helicase action is not required. -

USP7 Couples DNA Replication Termination to Mitotic Entry
bioRxiv preprint doi: https://doi.org/10.1101/305318; this version posted April 20, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. USP7 couples DNA replication termination to mitotic entry Antonio Galarreta1*, Emilio Lecona1*, Pablo Valledor1, Patricia Ubieto1,2, Vanesa Lafarga1, Julia Specks1 & Oscar Fernandez-Capetillo1,3 1Genomic Instability Group, Spanish National Cancer Research Centre (CNIO), Madrid 28029, Spain 2Current Address: DNA Replication Group, Spanish National Cancer Research Centre (CNIO), Madrid 28029, Spain 3Science for Life Laboratory, Division of Genome Biology, Department of Medical Biochemistry and Biophysics, Karolinska Institute, S-171 21 Stockholm, Sweden *Co-first authors Correspondence: E.L. ([email protected]) or O.F. ([email protected]) Lead Contact: Oscar Fernandez-Capetillo Spanish National Cancer Research Centre (CNIO) Melchor Fernandez Almagro, 3 Madrid 28029, Spain Tel.: +34.91.732.8000 Ext: 3480 Fax: +34.91.732.8028 Email: [email protected] KEYWORDS: USP7; CDK1; DNA REPLICATION; MITOSIS; S/M TRANSITION. bioRxiv preprint doi: https://doi.org/10.1101/305318; this version posted April 20, 2018. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. USP7 coordinates the S/M transition 2 SUMMARY To ensure a faithful segregation of chromosomes, DNA must be fully replicated before mitotic entry. However, how cells sense the completion of DNA replication and to what extent this is linked to the activation of the mitotic machinery remains poorly understood. We previously showed that USP7 is a replisome-associated deubiquitinase with an essential role in DNA replication. -

Catalytic Domain of Plasmid Pad1 Relaxase Trax Defines a Group Of
Catalytic domain of plasmid pAD1 relaxase TraX defines a group of relaxases related to restriction endonucleases María Victoria Franciaa,1, Don B. Clewellb,c, Fernando de la Cruzd, and Gabriel Moncaliánd aServicio de Microbiología, Hospital Universitario Marqués de Valdecilla e Instituto de Formación e Investigación Marqués de Valdecilla, Santander 39008, Spain; bDepartment of Biologic and Materials Sciences, University of Michigan School of Dentistry, Ann Arbor, MI 48109; cDepartment of Microbiology and Immunology, University of Michigan Medical School, Ann Arbor, MI 48109; and dDepartamento de Biología Molecular e Instituto de Biomedicina y Biotecnología de Cantabria, Universidad de Cantabria–Consejo Superior de Investigaciones Científicas–Sociedad para el Desarrollo Regional de Cantabria, Santander 39011, Spain Edited by Roy Curtiss III, Arizona State University, Tempe, AZ, and approved July 9, 2013 (received for review May 30, 2013) Plasmid pAD1 is a 60-kb conjugative element commonly found in known relaxases show two characteristic sequence motifs, motif I clinical isolates of Enterococcus faecalis. The relaxase TraX and the containing the catalytic Tyr residue (which covalently attaches to primary origin of transfer oriT2 are located close to each other and the 5′ end of the cleaved DNA) and motif III with the His-triad have been shown to be essential for conjugation. The oriT2 site essential for relaxase activity (facilitating the cleavage reaction contains a large inverted repeat (where the nic site is located) by activation of the catalytic Tyr). This His-triad has been used as adjacent to a series of short direct repeats. TraX does not show a relaxase diagnostic signature (12). fi any of the typical relaxase sequence motifs but is the prototype of Plasmid pAD1 relaxase, TraX, was identi ed (9) and shown to oriT2 a unique family of relaxases (MOB ). -

Teacher Notes Science Nspired
DNA Replication TEACHER NOTES SCIENCE NSPIRED Science Objectives In this lesson, students will • Explore how the structure of DNA supports its semi-conservative replication • Identify the name and function of several key enzymes in DNA replication • Recognize the function of Okazaki fragments in DNA replication Vocabulary TI-Nspire™ Technology Skills: • Double helix • Leading strand • Download a TI-Nspire • Lagging strand • Polymerase I • Polymerase III • Helicase document • Base pairs • Okazaki Fragments • Open a document • Primase • Move between pages • Open Directions Box About the Lesson • Explore ‘Hot Spots’ • Using three simulations, students will interact with DNA replication to explore semi-conservative replication and identify Tech Tips: specific enzymes and their roles in replication. Assessments are Make sure that students know embedded in the activity to engage discussion and gauge how to move between pages by learning. pressing /¡ (left arrow) and • As a result, students will: /¢ (right arrow). • Learn the basic functions of the following DNA replication enzymes: helicase, primase, ligase, polymerase I and III. Lesson Materials: • Learn how the double helix reproduces in a semi- Student Activity • DNA_Replication_Student.do conservative fashion c • Learn the role and function of Okazaki fragments in • DNA_Replication_Student.pd replication of the lagging strand. f TI-Nspire document TI-Nspire™ Navigator™ • DNA_Replication.tns Not required. If using Navigator: • Send out the DNA_Replication.tns file. • Monitor student progress -
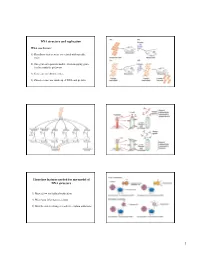
DNA Structure and Replication Three Key Features Needed for Any Model
DNA structure and replication What was known? 1) Hereditary factors were associated with specific traits 2) One-gene-one-protein model - from mapping genes for biosynthetic pathways 3) Genes are on chromosomes 4) Chromosomes are made up of DNA and protein Three key features needed for any model of DNA structure 1) Must allow for faithful replication 2) Must have information content 3) Must be able to change in order to explain mutations 1 Rosalind Franklin James Watson Francis Crick X-ray diffraction Maurice Wilkins Photograph of B-DNA Linus Pauling Nature 171, 737-738 (1953) © Macmillan Publishers Ltd. Molecular structure of Nucleic Acids WATSON, J. D. & CRICK, F. H. C. Features of the double helix A Structure for Deoxyribose Nucleic Acid 1) Two parallel strands We wish to suggest a structure for the salt of deoxyribose nucleic acid (D.N.A.). This structure has novel features which are of considerable biological interest………… 2) Bases held together by H-bonds 3) Phosphodiester backbone 4) The base is attached to the position 1 on the sugar 5) Base pair stack - provides Figure 1. This figure is purely diagrammatic. The two ribbons symbolize the two phophate-sugar chains, and the horizonal rods stability the pairs of bases holding the chains together. The vertical line marks the fibre axis. 6) Contains a major and …………….It has not escaped our notice that the specific pairing we minor groove have postulated immediately suggests a possible copying mechanism for the genetic material. Three key features needed for any model of DNA structure -

Processivity of DNA Polymerases: Two Mechanisms, One Goal Zvi Kelman1*, Jerard Hurwitz1 and Mike O’Donnell2
Minireview 121 Processivity of DNA polymerases: two mechanisms, one goal Zvi Kelman1*, Jerard Hurwitz1 and Mike O’Donnell2 Replicative DNA polymerases are highly processive Processive DNA synthesis by cellular replicases and the enzymes that polymerize thousands of nucleotides without bacteriophage T4 replicase dissociating from the DNA template. The recently Until recently, the only mechanism for high processivity determined structure of the Escherichia coli bacteriophage that was understood in detail was that utilized by cellular T7 DNA polymerase suggests a unique mechanism that replicases and the replicase of bacteriophage T4. This underlies processivity, and this mechanism may generalize mechanism involves a ring-shaped protein called a ‘DNA to other replicative polymerases. sliding clamp’ that encircles the DNA and tethers the polymerase catalytic unit to the DNA [3,4]. The three- Addresses: 1Department of Molecular Biology, Memorial Sloan- dimensional structures of several sliding clamps have been Kettering Cancer Center, 1275 York Avenue, New York, NY 10021, 2 determined: the eukaryotic proliferating cell nuclear USA and Laboratory of DNA Replication, Howard Hughes Medical β Institute, The Rockefeller University, 1230 York Avenue, New York, NY antigen (PCNA) [5,6]; the subunit of the prokaryotic 10021, USA. DNA polymerase III [7]; and the bacteriophage T4 gene 45 protein (gp45) (J Kuriyan, personal communication) *Corresponding author. (Figure 1). The overall structure of these clamps is very E-mail: [email protected] similar; the PCNA, β subunit and gp45 rings are super- Structure 15 February 1998, 6:121–125 imposable [8]. Each ring has similar dimensions and a http://biomednet.com/elecref/0969212600600121 central cavity large enough to accommodate duplex DNA (Figure 1). -

Polymerase Δ Deficiency Causes Syndromic Immunodeficiency with Replicative Stress
Polymerase δ deficiency causes syndromic immunodeficiency with replicative stress Cecilia Domínguez Conde, … , Mirjam van der Burg, Kaan Boztug J Clin Invest. 2019. https://doi.org/10.1172/JCI128903. Research Article Genetics Immunology Graphical abstract Find the latest version: https://jci.me/128903/pdf The Journal of Clinical Investigation RESEARCH ARTICLE Polymerase δ deficiency causes syndromic immunodeficiency with replicative stress Cecilia Domínguez Conde,1,2 Özlem Yüce Petronczki,1,2,3 Safa Baris,4,5 Katharina L. Willmann,1,2 Enrico Girardi,2 Elisabeth Salzer,1,2,3,6 Stefan Weitzer,7 Rico Chandra Ardy,1,2,3 Ana Krolo,1,2,3 Hanna Ijspeert,8 Ayca Kiykim,4,5 Elif Karakoc-Aydiner,4,5 Elisabeth Förster-Waldl,9 Leo Kager,6 Winfried F. Pickl,10 Giulio Superti-Furga,2,11 Javier Martínez,7 Joanna I. Loizou,2 Ahmet Ozen,4,5 Mirjam van der Burg,8 and Kaan Boztug1,2,3,6 1Ludwig Boltzmann Institute for Rare and Undiagnosed Diseases, 2CeMM Research Center for Molecular Medicine of the Austrian Academy of Sciences, and 3St. Anna Children’s Cancer Research Institute (CCRI), Vienna, Austria. 4Pediatric Allergy and Immunology, Marmara University, Faculty of Medicine, Istanbul, Turkey. 5Jeffrey Modell Diagnostic Center for Primary Immunodeficiency Diseases, Marmara University, Istanbul, Turkey. 6St. Anna Children’s Hospital, Department of Pediatrics and Adolescent Medicine, Vienna, Austria. 7Center for Medical Biochemistry, Medical University of Vienna, Vienna, Austria. 8Department of Pediatrics, Laboratory for Immunology, Leiden University Medical Centre, Leiden, Netherlands. 9Department of Neonatology, Pediatric Intensive Care and Neuropediatrics, Department of Pediatrics and Adolescent Medicine, 10Institute of Immunology, Center for Pathophysiology, Infectiology and Immunology, and 11Center for Physiology and Pharmacology, Medical University of Vienna, Vienna, Austria. -

The Obscure World of Integrative and Mobilizable Elements Gérard Guédon, Virginie Libante, Charles Coluzzi, Sophie Payot-Lacroix, Nathalie Leblond-Bourget
The obscure world of integrative and mobilizable elements Gérard Guédon, Virginie Libante, Charles Coluzzi, Sophie Payot-Lacroix, Nathalie Leblond-Bourget To cite this version: Gérard Guédon, Virginie Libante, Charles Coluzzi, Sophie Payot-Lacroix, Nathalie Leblond-Bourget. The obscure world of integrative and mobilizable elements: Highly widespread elements that pirate bacterial conjugative systems. Genes, MDPI, 2017, 8 (11), pp.337. 10.3390/genes8110337. hal- 01686871 HAL Id: hal-01686871 https://hal.archives-ouvertes.fr/hal-01686871 Submitted on 26 May 2020 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Distributed under a Creative Commons Attribution| 4.0 International License G C A T T A C G G C A T genes Review The Obscure World of Integrative and Mobilizable Elements, Highly Widespread Elements that Pirate Bacterial Conjugative Systems Gérard Guédon *, Virginie Libante, Charles Coluzzi, Sophie Payot and Nathalie Leblond-Bourget * ID DynAMic, Université de Lorraine, INRA, 54506 Vandœuvre-lès-Nancy, France; [email protected] (V.L.); [email protected] (C.C.); [email protected] (S.P.) * Correspondence: [email protected] (G.G.); [email protected] (N.L.-B.); Tel.: +33-037-274-5142 (G.G.); +33-037-274-5146 (N.L.-B.) Received: 12 October 2017; Accepted: 15 November 2017; Published: 22 November 2017 Abstract: Conjugation is a key mechanism of bacterial evolution that involves mobile genetic elements. -

Primer Release Is the Rate-Limiting Event in Lagging-Strand Synthesis
Primer release is the rate-limiting event in lagging- strand synthesis mediated by the T7 replisome Alfredo J. Hernandeza, Seung-Joo Leea, and Charles C. Richardsona,1 aDepartment of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, Boston, MA 02115 Contributed by Charles C. Richardson, April 18, 2016 (sent for review December 30, 2015; reviewed by Nicholas E. Dixon and I. Robert Lehman) DNA replication occurs semidiscontinuously due to the antiparallel but also for enabling the use of short oligoribonucleotides by T7 DNA strands and polarity of enzymatic DNA synthesis. Although DNA polymerase. Critically, the primase domain also fulfills two the leading strand is synthesized continuously, the lagging strand additional roles apart from primer synthesis: it prevents disso- is synthesized in small segments designated Okazaki fragments. ciation of the extremely short tetramer, stabilizing it with the Lagging-strand synthesis is a complex event requiring repeated template, and it secures it in the polymerase active site (10, 12). cycles of RNA primer synthesis, transfer to the lagging-strand Here we show that the rate-limiting step in initiation of Okazaki polymerase, and extension effected by cooperation between DNA fragments by the T7 replisome is primer release from the primase primase and the lagging-strand polymerase. We examined events domain of gp4. In the absence of gp2.5, an additional step, distinct controlling Okazaki fragment initiation using the bacteriophage from primer release, also limits primer extension. The presence of T7 replication system. Primer utilization by T7 DNA polymerase is gp2.5 promotes efficient primer formation and primer utilization. slower than primer formation. -

Virus World As an Evolutionary Network of Viruses and Capsidless Selfish Elements
Virus World as an Evolutionary Network of Viruses and Capsidless Selfish Elements Koonin, E. V., & Dolja, V. V. (2014). Virus World as an Evolutionary Network of Viruses and Capsidless Selfish Elements. Microbiology and Molecular Biology Reviews, 78(2), 278-303. doi:10.1128/MMBR.00049-13 10.1128/MMBR.00049-13 American Society for Microbiology Version of Record http://cdss.library.oregonstate.edu/sa-termsofuse Virus World as an Evolutionary Network of Viruses and Capsidless Selfish Elements Eugene V. Koonin,a Valerian V. Doljab National Center for Biotechnology Information, National Library of Medicine, Bethesda, Maryland, USAa; Department of Botany and Plant Pathology and Center for Genome Research and Biocomputing, Oregon State University, Corvallis, Oregon, USAb Downloaded from SUMMARY ..................................................................................................................................................278 INTRODUCTION ............................................................................................................................................278 PREVALENCE OF REPLICATION SYSTEM COMPONENTS COMPARED TO CAPSID PROTEINS AMONG VIRUS HALLMARK GENES.......................279 CLASSIFICATION OF VIRUSES BY REPLICATION-EXPRESSION STRATEGY: TYPICAL VIRUSES AND CAPSIDLESS FORMS ................................279 EVOLUTIONARY RELATIONSHIPS BETWEEN VIRUSES AND CAPSIDLESS VIRUS-LIKE GENETIC ELEMENTS ..............................................280 Capsidless Derivatives of Positive-Strand RNA Viruses....................................................................................................280 -

Junction Ribonuclease: an Activity in Okazaki Fragment Processing
Proc. Natl. Acad. Sci. USA Vol. 95, pp. 2244–2249, March 1998 Biochemistry Junction ribonuclease: An activity in Okazaki fragment processing RICHARD S. MURANTE,LEIGH A. HENRICKSEN, AND ROBERT A. BAMBARA* Department of Biochemistry and Biophysics, and Cancer Center, Box 712, University of Rochester School of Medicine and Dentistry, 601 Elmwood Avenue, Rochester, NY 14642 Communicated by I. Robert Lehman, Stanford University School of Medicine, Stanford, CA, December 29, 1997 (received for review September 15, 1997) ABSTRACT The initiator RNAs of mammalian Okazaki and removal of RNA primers. Reconstitution assays with both fragments are thought to be removed by RNase HI and the mouse cell and SV40 replication proteins require RNase HI for 5*-3* flap endonuclease (FEN1). Earlier evidence indicated the maturation of lagging strands in vitro (12, 13). that the cleavage site of RNase HI is 5* of the last ribonucle- Although cleavage of long RNAyDNA hybrids is random, otide at the RNA-DNA junction on an Okazaki substrate. In cleavage of certain substrates is clearly structure specific. Eder current work, highly purified calf RNase HI makes this exact et al. (16) found that when a series of ribonucleotides were cleavage in Okazaki fragments containing mismatches that followed by a 39 DNA segment and annealed to DNA to form distort the hybrid structure of the heteroduplex. Furthermore, a duplex, human RNase HI preferentially cleaved to leave a even fully unannealed Okazaki fragments were cleaved. DNA segment with a 59 monoribonucleotide. In reactions Clearly, the enzyme recognizes the transition from RNA to reconstituting Okazaki fragment processing, RNase HI from DNA on a single-stranded substrate and not the RNAyDNA calf thymus similarly cleaved the RNA-DNA strand 59 to the heteroduplex structure. -

The Bacterial Conjugation Protein Trwb Resembles Ring Helicases And
letters to nature metabolically labelled with BrdU (10 mM, 4 h), trypsinized, and ®xed with 70% ethanol. 17. Stampfer, M. R. et al. Gradual phenotypic conversion associated with immortalization of cultured Nuclei were isolated and stained with propidium iodide and FITC-conjugated anti-BrdU human mammary epithelial cells. Mol. Biol. Cell 8, 2391±2405 (1997). antibodies (Becton Dickinson, USA), as described7. Flow cytometry was performed on a 18. Karlseder, J., Broccoli, D., Dai, Y., Hardy, S. & de Lange, T. p53- and ATM-dependent apoptosis FACS Sorter (Becton Dickinson). All analysed events were gated to remove debris and induced by telomeres lacking TRF2. Science 283, 1321±1325 (1999). aggregates. 19. Artandi, S. E. et al. Telomere dysfunction promotes non-reciprocal translocations and epithelial cancers in mice. Nature 406, 641±645 (2000). Cell death assays 20. Chin, L. et al. p53 De®ciency rescues the adverse effects of telomere loss and cooperates with telomere dysfunction to accelerate carcinogenesis. Cell 97, 527±538 (1999). TUNEL assay for DNA fragmentation was done using an In Situ Cell Death Detection kit 21. Alcorta, D. A. et al. Involvement of the cyclin-dependent kinase inhibitor p16 (INK4A) in replicative (BMB), according to manufacturer's protocol. Alternatively, cells were stained with senescence of normal human ®broblasts. Proc. Natl Acad. Sci. USA 92, 13742±13747 (1996). Annexin-V-FLUOR (BMB) and propidium iodide, and analysed by ¯uorescence 22. Hara, E. et al. Regulation of p16CDKN2 expression and its implications for cell immortalization and microscopy. senescence. Mol. Cell. Biol. 16, 859±867 (1996). 23. Burbano, R. R. et al.