Lemons: Diversity and Relationships with Selected Citrus Genotypes As Measured with Nuclear Genome Markers
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Gin & Mixers Gins
GIN & MIXERS Some of our current favourites Malfy Grapefruit Mediterranean tonic, grapefruit 6.9 Hoxton Ginger ale, grapefruit 7.4 Boe Passion Fruit Lemonade, lime 5.8 King of Soho Mediterranean tonic, orange 7.4 Slingsby Gooseberry Tonic, lime 7.4 Silent Pool Tonic, orange 7.4 Tarquin’s Blackberry Lemonade, mint 6.3 Please ask us about our ‘Gin of the Week’ GINS 5th Gin Black Air Classic London Dry 5 Alkemist Muscat Grapes, Citrus, Rose 5.5 Audemus Pink Pepper Pink Pepper, Honey, Vanilla, Tonka Bean 5 Aviation Lavender, Anise, Sarsparilla 5 Beefeater 24 Green Tea, Sevilla Orange, Grapefruit 4.5 Beefeater Blood Orange Classic London Dry, Bittersweet Citrus Twist 4 Beefeater Peach & Raspberry Fresh Peach, Raspberry Tang 4 Beefeater Pink Strawberry Creamy Vanilla, Black Pepper, Soft Strawberry 4 Berliner Brandstifter Juniper, Fresh Blossoms, Elderflower 5.5 Bishops Lemongrass, Nasturtium Leaves, Lemon 5 Bitter Truth Juniper, Lemon, Caraway, Fennel 5 Black Tomato Black Tomatoes, Fresh Salt Water 5 Blackwoods Vintage Dry Juniper, Coriander, Lime 4.5 Bloom Honeysuckle, Chamomile, Pomelo 4.5 Blue Bottle Gorse Flowers, Nutmeg, Cubeb Pepper 5.5 Bobby’s Scheidam Cloves, Lemongrass, Cubeb Pepper 5 Boe Peach & Hibiscus Peach & Hibiscus Liqueur 4.5 Boe Violet Violet, Floral, Citrus 5 Boe Passion Fruit Passionfruit, Orange, Black Pepper 4.5 Bombay Sapphire Classic London Dry 4.5 Botanist Bog Myrtle, Mugwort, Meadowsweet 5 Brecon Botanicals Orange, Cinnamon, Cloves 4.5 Brecon Special Reserve Juniper, Coriander, Nutmeg, Cinnamon 4.5 Brockmans Blueberry, -

Meyer Lemon Concentrate.Indd
Meyer Lemon Concentrate Fresh Meyer Lemon is less acidic than the more common Eureka lemon. Its distinctive and complex citrus fl avor has hints of sweet lime, lemon and mandarin orange. Our Meyer Lemon Concentrate captures the fruit’s unique fl avor characteristics and perfumy aroma, with no added sugar or artifi cial Product Specifi cs ingredients. Ingredient List: Filtered water, Meyer lemon juice concentrate and natural lemon fl avor Meyer Lemon Pack Size: 6/30 oz. wide mouthed HDPE jars per case. Each jar attaches to a standard bar pour spout. Serving Size: 1 oz.. (28g) Servings per Container: 30 Brix: 18 - 20 Kosher: Amount Per Serving %Daily Value* Conversion: 1- 30 oz. = 0.85 kg Net Wt. Calories 20 1- 6/30 oz. case = 5.1 kg Net Wt. Total Fat 0g 0% Approx. fl . oz. per jar = 26 fl . oz. Sodium 0mg 0% Handling: Keep frozen. Product good for 7-10 days Total Carbohydrate 5g 2% thawed and refrigerated at 40° F and up to 24 months Sugars 1g frozen from manufactured date. Protein 0g Complimentary Flavors: Blueberry, white chocolate, Vitamin C 15% honey, almond Not a signifi cant source of calories from fat, saturated fat, trans fat, cholesterol, dietary fi ber, vitamin A, cal- Flavor Alternatives: Other high acid fruits like cium and iron. Passion Fruit Concentrate, Key Lime Concentrate and *Percent Daily Values are based on a 2,000 calorie diet. Blood Orange Concentrate Dilution Information Meyer Lemon Concentrate is stronger than straight Meyer lemon juice. Although it is mouth-puckering to taste, once blended with other ingredients it will not overpower a recipe and will remain true to the fl avor of fresh fruit. -

Chemical Variability of Peel and Leaf Essential Oils in the Citrus Subgenus Papeda (Swingle) and Relatives
Chemical variability of peel and leaf essential oils in the Citrus subgenus Papeda (Swingle) and relatives Clémentine Baccati, Marc Gibernau, Mathieu Paoli, Patrick Ollitrault, Félix Tomi, François Luro To cite this version: Clémentine Baccati, Marc Gibernau, Mathieu Paoli, Patrick Ollitrault, Félix Tomi, et al.. Chemical variability of peel and leaf essential oils in the Citrus subgenus Papeda (Swingle) and relatives. Plants, MDPI, 2021, 10 (6), pp.1117. 10.3390/plants10061117. hal-03262123 HAL Id: hal-03262123 https://hal.archives-ouvertes.fr/hal-03262123 Submitted on 16 Jun 2021 HAL is a multi-disciplinary open access L’archive ouverte pluridisciplinaire HAL, est archive for the deposit and dissemination of sci- destinée au dépôt et à la diffusion de documents entific research documents, whether they are pub- scientifiques de niveau recherche, publiés ou non, lished or not. The documents may come from émanant des établissements d’enseignement et de teaching and research institutions in France or recherche français ou étrangers, des laboratoires abroad, or from public or private research centers. publics ou privés. Distributed under a Creative Commons Attribution| 4.0 International License Chemical variability of peel and leaf essential oils in the Citrus subgenus Papeda (Swingle) and relatives Clémentine Baccati 1, Marc Gibernau 1, Mathieu Paoli 1, Patrick Ollitrault 2,3, Félix Tomi 1, * and François Luro 2 1 Université de Corse-CNRS, UMR 6134 SPE, Route des Sanguinaires, 20000 Ajaccio, France; [email protected] (C.B.) ; [email protected] (M.G.) ; [email protected] (M.P.) ; [email protected] (F.T.) 2 UMR AGAP Institut, Univ Montpellier, CIRAD, INRAE, Institut Agro – 20230, San Giuliano, France 3 CIRAD, UMR AGAP, F-20230 San Giuliano, France * Correspondence: [email protected]; tel.:+33-495-52-4122. -

Citrus Group 3-1.1
CPVO-TP/203/1 Date: 18/11/2004 EUROPEAN UNION COMMUNITY PLANT VARIETY OFFICE PROTOCOL FOR DISTINCTNESS, UNIFORMITY AND STABILITY TESTS Citrus L. – Group 3 LEMONS and LIMES UPOV Species Code: CITRU, CITRU_AUR, CITRU_LAT, CITRU_LIM Adopted on 18/11/2004 1 CPVO-TP/203/1 Date: 18/11/2004 I SUBJECT OF THE PROTOCOL The protocol describes the technical procedures to be followed in order to meet the Council Regulation 2100/94 on Community Plant Variety Rights. The technical procedures have been agreed by the Administrative Council and are based on general UPOV Document TG/1/3 and UPOV guideline TG/203/1 dated 09/04/2003 for the conduct of tests for Distinctness, Uniformity and Stability. This protocol applies for all varieties of the following group of the genus Citrus L. ( Rutaceae ), and their hybrids: LEMONS AND LIMES. See below for the list of species and their subgroups: Botanical taxon Sub- Common name group Citrus assamensis S. Dutta & S.C. Bhattach. LEM Citrus aurantiifolia (Christm.) Swingle SAL Mexican Lime Citrus aurata Risso LEM Citrus balotina Poit. & Turpin LEM Citrus bergamia Risso & Poit. SAL Citrus davaoensis (Wester) Tanaka SAL Citrus duttae Tanaka LEM Citrus excelsa Wester SAL Citrus hyalopulpa Tanaka SAL Citrus jambhiri Lush. LEM Rough Lemon (RLM) Citrus javanica Blume SAL Citrus karna Raf. LEM Citrus latifolia (Yu. Tanaka) Tanaka SAL Acid Lime (LAL) Citrus limetta Risso LEM Citrus limettioides Tanaka SAL Sweet Lime (SWL) Citrus limon (L.) Burm. f. LEM Lemon Citrus limon (L.) Burm. x C. aurantifolia HLL Lemonime (Christm.) Swing. Citrus limonia Osbeck LEM Citrus longilimon Tanaka LEM Citrus longispina Wester SAL Citrus lumia Risso & Poit. -

Citrus Limetta (Sweet Lemon, Mediterranean Sweet Lemon) Citrus Limetta Is a Small Tree with Lemon-Like Shape That Can Reach up to 8 M in Height
Citrus limetta (Sweet lemon, Mediterranean sweet lemon) Citrus limetta is a small tree with lemon-like shape that can reach up to 8 m in height. It has irregular branches, and relatively smooth, brownish-grey bark. It possesses numerous thorns. The leaves are more rounded and oval than an orange tree. Blossoms and new leaves are bright purple Landscape Information French Name: Limettier ﻟﻴﻤﻮﻥ ﺣﺎﻣﺾ :Arabic Name Pronounciation: SIT-rus lime-ET-ta Plant Type: Tree Origin: Asia Heat Zones: 9, 10, 11 Hardiness Zones: 10, 11, 12 Uses: Mass Planting, Container, Edible Size/Shape Tree Shape: Canopy Symmetry: Irregular Height at Maturity: 1.5 to 3 m Plant Image Spread at Maturity: 3 to 5 meters, 5 to 8 meters Citrus limetta (Sweet lemon, Mediterranean sweet lemon) Botanical Description Foliage Leaf Arrangement: Alternate Leaf Venation: Pinnate Leaf Persistance: Evergreen Leaf Type: Simple Leaf Blade: 5 - 10 cm Leaf Margins: Crenate Leaf Textures: Glossy Leaf Scent: Pleasant Color(growing season): Green Color(changing season): Green Flower Fruit Image Flower Showiness: True Flower Size Range: 1.5 - 3 Flower Scent: Pleasant Flower Color: White Seasons: Spring Trunk Trunk Esthetic Values: Smooth, Spines Fruit Fruit Type: Hesperidium Fruit Showiness: True Fruit Colors: Yellow Seasons: Spring Citrus limetta (Sweet lemon, Mediterranean sweet lemon) Horticulture Management Tolerance Frost Tolerant: No Heat Tolerant: No Drought Tolerant: Yes Salt Tolerance: Moderate Requirements Soil Requirements: Loam, Sand Soil Ph Requirements: Acidic Water Requirements: Moderate Light Requirements: Management Edible Parts: Other Image Plant Propagations: Grafting. -

Citrus Problems – Sprouting Rootstock
CITRUS PROBLEMS – SPROUTING ROOTSTOCK The citrus trees you purchase at the nursery have all been grafted. That is, a desirable, named citrus variety, such as Owari satsuma or Meyer lemon, is grafted onto a rootstock that is a completely different type of citrus. Trifoliata orange (also called sour orange) is often used as the rootstock. The point where the graft was made (called the graft union) will generally appear as a swollen point or crook in the lower part of a trunk. When you purchase a young citrus tree, look for and find the graft union. Everything above the graft union is the desirable citrus tree – the satsuma, lemon, kumquat, orange or grapefruit – called the scion. Everything below the graft union is something else entirely – either trifoliata orange (Poncirus trifoliata Rubidoux) or Swingle citrumello – called the rootstock. The purpose of the rootstock is to provide a strong, vigorous root system that will produce a robust growing, productive tree. The advantage of the trifoliata root stock is that is also imparts increased cold hardiness to the upper part of the tree Once you have located the graft union on the trunk, you must never allow any shoots to sprout and grow from below the graft union. These shoots are called “suckers.” If you let these vigorous suckers grow, you are allowing something that is not your desirable citrus variety to grow. When a citrus tree produces atypical fruit, it generally means the rootstock has been allowed to sprout and grow. The trifoliata rootstock produces poor quality, seedy, sour, round yellow fruit. -

Citrus from Seed?
Which citrus fruits will come true to type Orogrande, Tomatera, Fina, Nour, Hernandina, Clementard.) from seed? Ellendale Tom McClendon writes in Hardy Citrus Encore for the South East: Fortune Fremont (50% monoembryonic) “Most common citrus such as oranges, Temple grapefruit, lemons and most mandarins Ugli Umatilla are polyembryonic and will come true to Wilking type. Because most citrus have this trait, Highly polyembryonic citrus types : will mostly hybridization can be very difficult to produce nucellar polyembryonic seeds that will grow true to type. achieve…. This unique characteristic Citrus × aurantiifolia Mexican lime (Key lime, West allows amateurs to grow citrus from seed, Indian lime) something you can’t do with, say, Citrus × insitorum (×Citroncirus webberii) Citranges, such as Rusk, Troyer etc. apples.” [12*] Citrus × jambhiri ‘Rough lemon’, ‘Rangpur’ lime, ‘Otaheite’ lime Monoembryonic (don’t come true) Citrus × limettioides Palestine lime (Indian sweet lime) Citrus × microcarpa ‘Calamondin’ Meyer Lemon Citrus × paradisi Grapefruit (Marsh, Star Ruby, Nagami Kumquat Redblush, Chironja, Smooth Flat Seville) Marumi Kumquat Citrus × sinensis Sweet oranges (Blonde, navel and Pummelos blood oranges) Temple Tangor Citrus amblycarpa 'Nasnaran' mandarin Clementine Mandarin Citrus depressa ‘Shekwasha’ mandarin Citrus karna ‘Karna’, ‘Khatta’ Poncirus Trifoliata Citrus kinokuni ‘Kishu mandarin’ Citrus lycopersicaeformis ‘Kokni’ or ‘Monkey mandarin’ Polyembryonic (come true) Citrus macrophylla ‘Alemow’ Most Oranges Citrus reshni ‘Cleopatra’ mandarin Changshou Kumquat Citrus sunki (Citrus reticulata var. austera) Sour mandarin Meiwa Kumquat (mostly polyembryonic) Citrus trifoliata (Poncirus trifoliata) Trifoliate orange Most Satsumas and Tangerines The following mandarin varieties are polyembryonic: Most Lemons Dancy Most Limes Emperor Grapefruits Empress Tangelos Fairchild Kinnow Highly monoembryonic citrus types: Mediterranean (Avana, Tardivo di Ciaculli) Will produce zygotic monoembryonic seeds that will not Naartje come true to type. -
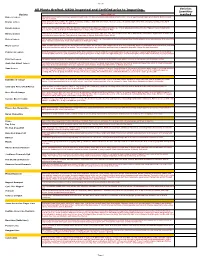
2019 Full Provisional List
Sheet1 All Plants Grafted. USDA inspected and Certified prior to Importing. Varieties Quantities Variety Description required Baboon Lemon A Brazilian lemon with very intense yellow rind and flesh. The flavour is acidic with almost a hint of lime. Tree is vigorous with large green leaves. Both tree and fruit are beautiful. Bearss Lemon 1952. Fruit closely resembles the Lisbon. Very juicy and has a high rind oil content. The leaves are a beautiful purple when first emerging, turning a nice dark green. Fruit is ready from June to December. Eureka Lemon Fruit is very juicy and highly acidic. The Eureka originated in Los Angeles, California and is one of their principal varieties. It is the "typical" lemon found in the grocery stores, nice yellow colour with typical lemon shape. Harvested November to May Harvey Lemon 1948.Having survived the disastrous deep freezes in Florida during the ’60’s and ’70’s. this varieties is known to withstand cold weather. Typical lemon shape and tart, juicy true lemon flavour. Fruit ripens in September to March. Self fertile. Zones 8A-10. Lisbon Lemon Fruit is very juicy and acic. The leaves are dense and tree is very vigorous. This Lisbon is more cold tolerant than the Eureka and is more productive. It is one of the major varieties in California. Fruit is harvested from February to May. Meyer Lemon 1908. Considered ever-bearing, the blooms are very aromatic. It is a lemon and orange hybrid. It is very cold hardy. Fruit is round with a thin rind. Fruit is juicy and has a very nice flavour, with a low acidity. -

FEMA GRAS Assessment of Natural Flavor Complexes Citrus-Derived
Food and Chemical Toxicology 124 (2019) 192–218 Contents lists available at ScienceDirect Food and Chemical Toxicology journal homepage: www.elsevier.com/locate/foodchemtox FEMA GRAS assessment of natural flavor complexes: Citrus-derived T flavoring ingredients Samuel M. Cohena, Gerhard Eisenbrandb, Shoji Fukushimac, Nigel J. Gooderhamd, F. Peter Guengeriche, Stephen S. Hechtf, Ivonne M.C.M. Rietjensg, Maria Bastakih, ∗ Jeanne M. Davidsenh, Christie L. Harmanh, Margaret McGowenh, Sean V. Taylori, a Havlik-Wall Professor of Oncology, Dept. of Pathology and Microbiology, University of Nebraska Medical Center, 983135 Nebraska Medical Center, Omaha, NE, 68198- 3135, USA b Food Chemistry & Toxicology, Kühler Grund 48/1, 69126 Heidelberg, Germany c Japan Bioassay Research Center, 2445 Hirasawa, Hadano, Kanagawa, 257-0015, Japan d Dept. of Surgery and Cancer, Imperial College London, Sir Alexander Fleming Building, London, SW7 2AZ, United Kingdom e Dept. of Biochemistry, Vanderbilt University School of Medicine, Nashville, TN, 37232-0146, USA f Masonic Cancer Center, Dept. of Laboratory Medicine and Pathology, University of Minnesota, Cancer and Cardiovascular Research Building, 2231 6th St. SE, Minneapolis, MN, 55455, USA g Division of Toxicology, Wageningen University, Stippeneng 4, 6708 WE, Wageningen, the Netherlands h Flavor and Extract Manufacturers Association, 1101 17th Street, NW Suite 700, Washington, DC, 20036, USA i Scientific Secretary to the FEMA Expert Panel, 1101 17th Street, NW Suite 700, Washington, DC,20036,USA ARTICLE INFO ABSTRACT Keywords: In 2015, the Expert Panel of the Flavor and Extract Manufacturers Association (FEMA) initiated a re-evaluation Citrus of the safety of over 250 natural flavor complexes (NFCs) used as flavoring ingredients. This publication isthe Natural flavor complex first in a series and summarizes the evaluation of54 Citrus-derived NFCs using the procedure outlined in Smith Botanical et al. -

Survey of Phenolic Compounds Produced in Citrus
USDA ??:-Z7 S rveyof Phenolic United States Department of Agriculture C mpounds Produced IliIIiI Agricultural Research In Citrus Service Technical Bulletin Number 1856 December 1998 United States Department of Agriculture Survey of Phenolic Compounds Agricultural Produced in Citrus Research Service Mark Berhow, Brent Tisserat, Katherine Kanes, and Carl Vandercook Technical Bulletin Number 1856 December 1998 This research project was conducted at USDA, Agricultural Research Service, Fruit and Vegetable Chem istry laboratory, Pasadena, California, where Berhow was a research chemist, TIsserat was a research geneticist, Kanes was a research associate, and Vandercook, now retired, was a research chemist. Berhow and Tisserat now work at the USDA-ARS National Center for AgriCUltural Utilization Research, Peoria, Illinois, where Berhow is a research chemist and Tisserat is a research geneticist. Abstract Berhow, M., B. Tisserat, K. Kanes, and C. Vandercook. 1998. Survey of Mention of trade names or companies in this publication is solely for the Phenolic Compounds Produced in Citrus. U.S. Department ofAgriculture, purpose of providing specific information and does not imply recommenda Agricultural Research Service, Technical Bulletin No. 1856, 158 pp. tion or endorsement by the U. S. Department ofAgriculture over others not mentioned. A survey of phenolic compounds, especially flavanones and flavone and flavonol compounds, using high pressure liquid chromatography was While supplies last, single copies of this publication may be obtained at no performed in Rutaceae, subfamily Aurantioideae, representing 5 genera, cost from- 35 species, and 114 cultivars. The average number of peaks, or phenolic USDA, ARS, National Center for Agricultural Utilization Research compounds, occurring in citrus leaf, flavedo, albedo, and juice vesicles 1815 North University Street were 21, 17, 15, and 9.3, respectively. -

High Biological Value Compounds Extraction from Citrus Waste with Non-Conventional Methods
foods Review High Biological Value Compounds Extraction from Citrus Waste with Non-Conventional Methods Mayra Anticona, Jesus Blesa , Ana Frigola and Maria Jose Esteve * Nutrition and Food Chemistry, University of Valencia, Avda., Vicent Andrés Estellés, s/n., 46100 Burjassot, Spain; [email protected] (M.A.); [email protected] (J.B.); [email protected] (A.F.) * Correspondence: [email protected]; Tel.: +34-963544913 Received: 27 April 2020; Accepted: 15 June 2020; Published: 20 June 2020 Abstract: Citrus fruits are extensively grown and much consumed around the world. Eighteen percent of total citrus cultivars are destined for industrial processes, and as a consequence, large amounts of waste are generated. Citrus waste is a potential source of high biological value compounds, which can be used in the food, pharmaceutical, and cosmetic industries but whose final disposal may pose a problem due to economic and environmental factors. At the same time, the emerging need to reduce the environmental impact of citrus waste and its responsible management has increased. For these reasons, the study of the use of non-conventional methods to extract high biological value compounds such as carotenoids, polyphenols, essential oils, and pectins from this type of waste has become more urgent in recent years. In this review, the effectiveness of technologies such as ultrasound assisted extraction, microwave assisted extraction, supercritical fluid extraction, pressurized water extraction, pulsed electric field, high-voltage electric discharges, and high hydrostatic pressures is described and assessed. A wide range of information concerning the principal non-conventional methods employed to obtain high-biological-value compounds from citrus waste as well as the most influencing factors about each technology are considered. -

Extraction of Pectin from Citrus Fruit Peel and Its Utilization in Preparation of Jelly
International Journal of Engineering Research & Technology (IJERT) ISSN: 2278-0181 Vol. 3 Issue 5, May - 2014 Extraction of Pectin from Citrus Fruit Peel and Its Utilization in Preparation of Jelly 1 W. Elizabeth Devi R N Shukla2 K L Bala3 A Kumar4 A A Mishra5 M.Tech in Food Technology (Food Process Engineering), K C Yadav6 Department of Food Process Engineering 2,3,4,5,6 Vaugh School of Agriculture Engineering, SHIATS Deemed Assistant Professor University Department of Food Process Engineering P.O-Naini, Allahabad, U.P-211007 Vaugh School of Agriculture Engineering, SHIATS Deemed University P.O-Naini, Allahabad, U.P-211007, India Abstract— The present study was focused on the potential of The term pectin was first described and isolated by Henry citrus peel as a source of pectin. Pectin was extracted from Sweet Braconnot in 1825 (Braconnot, 1825). Pectin is a lemon (mosambi) peel powder using two different acids (citric and polysaccharide, naturally occurring substance present in all plant nitric) and at three different temperatures, time and pH viz (60, 70 tissue. Pectin exists in varying amounts in fruit cell walls and & 80°C), (30,45 & 60 min),(1.5,2 & 2.5pH) respectively. Pectin has important nutritional and technological properties (Knox yields varied from 21.4% to 76.0% and 17.4% to 46.4% extracted by using citric acid and nitric acid respectively. The best extraction 2002). In the cell walls they serve as one of the main agents condition was found to be higher in yield by using citric acid at cementing the cellulose fibrils and may be linked covalently to 80°C, 60min, 1.5pH.