View of This Evidence with Insufficient Symptoms Or Fatigue (ISF)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Machine-Learning and Chemicogenomics Approach Defi Nes and Predicts Cross-Talk of Hippo and MAPK Pathways
Published OnlineFirst November 18, 2020; DOI: 10.1158/2159-8290.CD-20-0706 RESEARCH ARTICLE Machine -Learning and Chemicogenomics Approach Defi nes and Predicts Cross-Talk of Hippo and MAPK Pathways Trang H. Pham 1 , Thijs J. Hagenbeek 1 , Ho-June Lee 1 , Jason Li 2 , Christopher M. Rose 3 , Eva Lin 1 , Mamie Yu 1 , Scott E. Martin1 , Robert Piskol 2 , Jennifer A. Lacap 4 , Deepak Sampath 4 , Victoria C. Pham 3 , Zora Modrusan 5 , Jennie R. Lill3 , Christiaan Klijn 2 , Shiva Malek 1 , Matthew T. Chang 2 , and Anwesha Dey 1 ABSTRACT Hippo pathway dysregulation occurs in multiple cancers through genetic and non- genetic alterations, resulting in translocation of YAP to the nucleus and activation of the TEAD family of transcription factors. Unlike other oncogenic pathways such as RAS, defi ning tumors that are Hippo pathway–dependent is far more complex due to the lack of hotspot genetic alterations. Here, we developed a machine-learning framework to identify a robust, cancer type–agnostic gene expression signature to quantitate Hippo pathway activity and cross-talk as well as predict YAP/TEAD dependency across cancers. Further, through chemical genetic interaction screens and multiomics analyses, we discover a direct interaction between MAPK signaling and TEAD stability such that knockdown of YAP combined with MEK inhibition results in robust inhibition of tumor cell growth in Hippo dysregulated tumors. This multifaceted approach underscores how computational models combined with experimental studies can inform precision medicine approaches including predictive diagnostics and combination strategies. SIGNIFICANCE: An integrated chemicogenomics strategy was developed to identify a lineage- independent signature for the Hippo pathway in cancers. -

An Extracellular Site on Tetraspanin CD151 Determines Α3 and Α6
JCBArticle An extracellular site on tetraspanin CD151 determines ␣3 and ␣6 integrin–dependent cellular morphology Alexander R. Kazarov, Xiuwei Yang, Christopher S. Stipp, Bantoo Sehgal, and Martin E. Hemler Dana-Farber Cancer Institute and Department of Pathology, Harvard Medical School, Boston, MA 02115 he ␣31 integrin shows strong, stoichiometric, direct Brij 96–resistant) were independent of the QRD/TS151r lateral association with the tetraspanin CD151. As site, occurred late in biosynthesis, and involved mature T shown here, an extracellular CD151 site (QRD194–196) integrin subunits. Presence of the CD151–QRD194–196→INF is required for strong (i.e., Triton X-100–resistant) ␣31 mutant disrupted ␣3 and ␣6 integrin–dependent formation association and for maintenance of a key CD151 epitope of a network of cellular cables by Cos7 or NIH3T3 cells on (defined by monoclonal antibody TS151r) that is blocked basement membrane Matrigel and markedly altered cell upon ␣3 integrin association. Strong CD151 association spreading. These results provide definitive evidence that with integrin ␣61 also required the QRD194–196 site and strong lateral CD151–integrin association is functionally masked the TS151r epitope. For both ␣3 and ␣6 integrins, important, identify CD151 as a key player during ␣3 and strong QRD/TS151r-dependent CD151 association occurred ␣6 integrin–dependent matrix remodeling and cell spreading, early in biosynthesis and involved ␣ subunit precursor and support a model of CD151 as a transmembrane linker forms. In contrast, weaker associations of CD151 with itself, between extracellular integrin domains and intracellular integrins, or other tetraspanins (Triton X-100–sensitive but cytoskeleton/signaling molecules. -

4-6 Weeks Old Female C57BL/6 Mice Obtained from Jackson Labs Were Used for Cell Isolation
Methods Mice: 4-6 weeks old female C57BL/6 mice obtained from Jackson labs were used for cell isolation. Female Foxp3-IRES-GFP reporter mice (1), backcrossed to B6/C57 background for 10 generations, were used for the isolation of naïve CD4 and naïve CD8 cells for the RNAseq experiments. The mice were housed in pathogen-free animal facility in the La Jolla Institute for Allergy and Immunology and were used according to protocols approved by the Institutional Animal Care and use Committee. Preparation of cells: Subsets of thymocytes were isolated by cell sorting as previously described (2), after cell surface staining using CD4 (GK1.5), CD8 (53-6.7), CD3ε (145- 2C11), CD24 (M1/69) (all from Biolegend). DP cells: CD4+CD8 int/hi; CD4 SP cells: CD4CD3 hi, CD24 int/lo; CD8 SP cells: CD8 int/hi CD4 CD3 hi, CD24 int/lo (Fig S2). Peripheral subsets were isolated after pooling spleen and lymph nodes. T cells were enriched by negative isolation using Dynabeads (Dynabeads untouched mouse T cells, 11413D, Invitrogen). After surface staining for CD4 (GK1.5), CD8 (53-6.7), CD62L (MEL-14), CD25 (PC61) and CD44 (IM7), naïve CD4+CD62L hiCD25-CD44lo and naïve CD8+CD62L hiCD25-CD44lo were obtained by sorting (BD FACS Aria). Additionally, for the RNAseq experiments, CD4 and CD8 naïve cells were isolated by sorting T cells from the Foxp3- IRES-GFP mice: CD4+CD62LhiCD25–CD44lo GFP(FOXP3)– and CD8+CD62LhiCD25– CD44lo GFP(FOXP3)– (antibodies were from Biolegend). In some cases, naïve CD4 cells were cultured in vitro under Th1 or Th2 polarizing conditions (3, 4). -

Page 1 Supplemental Table I Upin WTVG Upin Humlowfb FAR1
Supplemental Table I UPinWTvG UPinHuMlowFB overlap FAR1 CD209 PLEKHO2 B2M IL27 GCLC CALR HMGN2P46 ME1 XPO1 CLEC1A NEK6 PDIA3 HSD17B14 TMEM106A SERPINH1 NSUN7 KCNJ15 PLEKHO2 SEMA6B SH3PXD2B HSPD1 BAALC RHOU SYVN1 CEACAM4 TGFBI DNAJB9 KATNAL2 CTSZ SLC25A19 CCDC175 SULF2 MHCII CARD14 P2RY6 MDN1 TDO2 CD74 GCLC FCAR HLA-DQB1 PTPN2 GLDN CD14 CHORDC1 MMP7 CSF2RB LOX CLEC5A MRC1 STIP1 ZMYND15 ITGAX BPIFB1 ITLN1 SLAMF7 ME1 DKK2 CD84 PDIA6 FAM124A FCGR2A HYOU1 F3 CLEC10A NLRC5 DZIP1L IFI30 NEK6 CECR6 CLEC4A SLC39A14 NDP CLEC7A TMEM106AFCGR1A TFEC KCNJ15 CCL7 C1QC SH3PXD2B CRABP2 C1QA RHOU PIPOX FOLR2 LRP2 CCL2 CH25H MVD VSIG4 C1QB TGFBI LINC01010 SIGLEC1 NFKB2 DPRXP4 CTSS C5 FAM20A CCR1 HSP90B1 ANKRD29 SLAMF8 ALDH18A1 OCSTAMP MS4A7 EDEM1 TGM2 HK3 CTSZ TM4SF19 CXCL10 C6 TRPV4 CTSK AACS CCL8 MSR1 PPA1 CCL1 STEAP4 PIK3R5 KCNE1 CXCL9 RASAL3 SLC9A7P1 MS4A6A DOCK11 CHI3L1 TIMP1 BHLHE40 LOC731424 CD209 HCLS1 DCSTAMP CCL7 FASN MSR1 PDCD1LG2 PDIA4 IL31RA CCL2 ITK CXCL3 CXCL2 CRELD2 TREM2 C15orf48 SFTPD MGST1 GPR84 BCL3 METTL7B CXCL5 SULF2 TMEM86A OCSTAMP CYP51A1 A2M SERPINA1 CREB3L1 AQP9 MMP12 DUSP2 NUPR1 CCL8 ADAM8 FHAD1 CCL24 P2RY6 YPEL4 FBP1 KCNAB2 FBP1 NA NFKBIE LOC100506585 NA FSCN1 CXCL16 NA MANF RAB13 NA SLC5A3 LOC391322 NA CTSC IL8 NA COTL1 MS4A4A NA HSPA5 SERPING1 NA MUC5B PLA2G4C NA CD74 CA12 NA HLA-DQB1 GBP1P1 NA SLC7A2 C11orf45 NA FABP5 ACVRL1 NA CIITA SPP1 NA RAB3IL1 TLN2 NA HSPE1 NDRG2 NA SCD C15orf48 NA ITIH4 KCNJ15 NA SERPINA3 MEIS3P1 NA LAG3 IL1RN NA FOXM1 HNMT NA CD14 CYP27B1 NA RRM2 CDCP1 NA ABCD2 FOLR2 NA FCRL2 ECM1 NA PDE3B ADAMDEC1 -

PRODUCT SPECIFICATION Anti-TSPAN3
Anti-TSPAN3 Product Datasheet Polyclonal Antibody PRODUCT SPECIFICATION Product Name Anti-TSPAN3 Product Number HPA015996 Gene Description tetraspanin 3 Clonality Polyclonal Isotype IgG Host Rabbit Antigen Sequence Recombinant Protein Epitope Signature Tag (PrEST) antigen sequence: NGTNPDAASRAIDYVQRQLHCCGIHNYSDWENTDWFKETKNQSVPLSCCR ETASNCNGSLAHPSDLYAEGCEALVVKKLQEI Purification Method Affinity purified using the PrEST antigen as affinity ligand Verified Species Human Reactivity Recommended IHC (Immunohistochemistry) Applications - Antibody dilution: 1:20 - 1:50 - Retrieval method: HIER pH6 ICC-IF (Immunofluorescence) - Fixation/Permeabilization: PFA/Triton X-100 - Working concentration: 0.25-2 µg/ml Characterization Data Available at atlasantibodies.com/products/HPA015996 Buffer 40% glycerol and PBS (pH 7.2). 0.02% sodium azide is added as preservative. Concentration Lot dependent Storage Store at +4°C for short term storage. Long time storage is recommended at -20°C. Notes Gently mix before use. Optimal concentrations and conditions for each application should be determined by the user. For protocols, additional product information, such as images and references, see atlasantibodies.com. Product of Sweden. For research use only. Not intended for pharmaceutical development, diagnostic, therapeutic or any in vivo use. No products from Atlas Antibodies may be resold, modified for resale or used to manufacture commercial products without prior written approval from Atlas Antibodies AB. Warranty: The products supplied by Atlas Antibodies are warranted to meet stated product specifications and to conform to label descriptions when used and stored properly. Unless otherwise stated, this warranty is limited to one year from date of sales for products used, handled and stored according to Atlas Antibodies AB's instructions. Atlas Antibodies AB's sole liability is limited to replacement of the product or refund of the purchase price. -

Complexes of Tetraspanins with Integrins: More Than Meets the Eye
COMMENTARY 4143 Complexes of tetraspanins with integrins: more than meets the eye Fedor Berditchevski CRC Institute for Cancer Studies, The University of Birmingham, Edgbaston, Birmingham, B15 2TA, UK Author for correspondence (e-mail: [email protected]) Journal of Cell Science 114, 4143-4151 (2001) © The Company of Biologists Ltd Summary The transmembrane proteins of the tetraspanin membrane, integrin-tetraspanin signalling complexes superfamily are implicated in a diverse range of biological are partitioned into specific microdomains proximal to phenomena, including cell motility, metastasis, cell cholesterol-rich lipid rafts. A substantial fraction of proliferation and differentiation. The tetraspanins are tetraspanins colocalise with integrins in various associated with adhesion receptors of the integrin family intracellular vesicular compartments. It is proposed that and regulate integrin-dependent cell migration. In cells tetraspanins can influence cell migration by one of the attached to the extracellular matrix, the integrin- following mechanisms: (1) modulation of integrin tetraspanin adhesion complexes are clustered into a distinct signalling; (2) compartmentalisation of integrins on the cell type of adhesion structure at the cell periphery. Various surface; or (3) direction of intracellular trafficking and tetraspanins are associated with phosphatidylinositol 4- recycling of integrins. kinase and protein kinase C isoforms, and they may facilitate assembly of signalling complexes by tethering these enzymes to integrin heterodimers. At the plasma Key words: Tetraspanin, Integrin, Migration, Signalling Introduction relatively well conserved among tetraspanins (Fig. 1). A Tetraspanins (also referred to as tetraspans or TM4SF proteins) combination of all the above features distinguishes tetraspanins are a family of widely expressed four-transmembrane-domain from a diverse group of proteins that have four transmembrane proteins. -

Downloaded 18 July 2014 with a 1% False Discovery Rate (FDR)
UC Berkeley UC Berkeley Electronic Theses and Dissertations Title Chemical glycoproteomics for identification and discovery of glycoprotein alterations in human cancer Permalink https://escholarship.org/uc/item/0t47b9ws Author Spiciarich, David Publication Date 2017 Peer reviewed|Thesis/dissertation eScholarship.org Powered by the California Digital Library University of California Chemical glycoproteomics for identification and discovery of glycoprotein alterations in human cancer by David Spiciarich A dissertation submitted in partial satisfaction of the requirements for the degree Doctor of Philosophy in Chemistry in the Graduate Division of the University of California, Berkeley Committee in charge: Professor Carolyn R. Bertozzi, Co-Chair Professor David E. Wemmer, Co-Chair Professor Matthew B. Francis Professor Amy E. Herr Fall 2017 Chemical glycoproteomics for identification and discovery of glycoprotein alterations in human cancer © 2017 by David Spiciarich Abstract Chemical glycoproteomics for identification and discovery of glycoprotein alterations in human cancer by David Spiciarich Doctor of Philosophy in Chemistry University of California, Berkeley Professor Carolyn R. Bertozzi, Co-Chair Professor David E. Wemmer, Co-Chair Changes in glycosylation have long been appreciated to be part of the cancer phenotype; sialylated glycans are found at elevated levels on many types of cancer and have been implicated in disease progression. However, the specific glycoproteins that contribute to cell surface sialylation are not well characterized, specifically in bona fide human cancer. Metabolic and bioorthogonal labeling methods have previously enabled enrichment and identification of sialoglycoproteins from cultured cells and model organisms. The goal of this work was to develop technologies that can be used for detecting changes in glycoproteins in clinical models of human cancer. -

Human Induced Pluripotent Stem Cell–Derived Podocytes Mature Into Vascularized Glomeruli Upon Experimental Transplantation
BASIC RESEARCH www.jasn.org Human Induced Pluripotent Stem Cell–Derived Podocytes Mature into Vascularized Glomeruli upon Experimental Transplantation † Sazia Sharmin,* Atsuhiro Taguchi,* Yusuke Kaku,* Yasuhiro Yoshimura,* Tomoko Ohmori,* ‡ † ‡ Tetsushi Sakuma, Masashi Mukoyama, Takashi Yamamoto, Hidetake Kurihara,§ and | Ryuichi Nishinakamura* *Department of Kidney Development, Institute of Molecular Embryology and Genetics, and †Department of Nephrology, Faculty of Life Sciences, Kumamoto University, Kumamoto, Japan; ‡Department of Mathematical and Life Sciences, Graduate School of Science, Hiroshima University, Hiroshima, Japan; §Division of Anatomy, Juntendo University School of Medicine, Tokyo, Japan; and |Japan Science and Technology Agency, CREST, Kumamoto, Japan ABSTRACT Glomerular podocytes express proteins, such as nephrin, that constitute the slit diaphragm, thereby contributing to the filtration process in the kidney. Glomerular development has been analyzed mainly in mice, whereas analysis of human kidney development has been minimal because of limited access to embryonic kidneys. We previously reported the induction of three-dimensional primordial glomeruli from human induced pluripotent stem (iPS) cells. Here, using transcription activator–like effector nuclease-mediated homologous recombination, we generated human iPS cell lines that express green fluorescent protein (GFP) in the NPHS1 locus, which encodes nephrin, and we show that GFP expression facilitated accurate visualization of nephrin-positive podocyte formation in -

Tetraspanin 3 (TSPAN3) Organism: Homo Sapiens (Human) Instruction Manual for in VITRO USE and RESEARCH USE ONLY NOT for USE in DIAGNOSTIC OR THERAPEUTIC PROCEDURES
P92816Hu01 Tetraspanin 3 (TSPAN3) Organism: Homo sapiens (Human) Instruction manual FOR IN VITRO USE AND RESEARCH USE ONLY NOT FOR USE IN DIAGNOSTIC OR THERAPEUTIC PROCEDURES 4th Edition (Revised in August, 2012) [ DESCRIPTION ] Protein Names: Tetraspanin 3 Synonyms: TSPAN3, TM4SF8 Species: Human Size: 100µg Source: Escherichia coli-derived Subcellular Location: Membrane; Multi-pass membrane protein. [ PROPERTIES ] Residues: Tyr107~Ile213 (Accession # O60637), with two N-terminal Tags, His-tag and S-tag. Grade & Purity: >95%, 18 kDa as determined by SDS-PAGE reducing conditions. Formulation: Supplied as liquid form in Phosphate buffered saline(PBS), pH 7.4. Endotoxin Level: <1.0 EU per 1μg (determined by the LAL method). Applications: SDS-PAGE; WB; ELISA; IP. (May be suitable for use in other assays to be determined by the end user.) Predicted Molecular Mass: 17.7 kDa Predicted isoelectric point: 6.0 [ PREPARATION ] Reconstitute in sterile PBS, pH7.2-pH7.4. [ STORAGE AND STABILITY ] Storage: Avoid repeated freeze/thaw cycles. Store at 2-8oC for one month. Aliquot and store at -80oC for 12 months. Stability Test: The thermal stability is described by the loss rate of the target protein. The loss rate was determined by accelerated thermal degradation test, that is, incubate the protein at 37oC for 48h, and no obvious degradation and precipitation were observed. (Referring from China Biological Products Standard, which was calculated by the Arrhenius equation.) The loss of this protein is less than 5% within the expiration date under appropriate storage condition. [ SEQUENCES ] The target protein is fused with two N-terminal Tags, His-tag and S-tag, its sequence is listed below. -
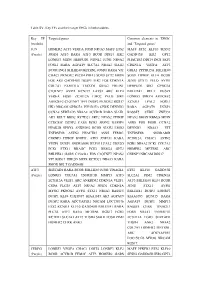
Targeted Genes Common Elements In
Table SV. Key TFs and their target DEGs in hub modules. Key TF Targeted genes Common elements in ‘DEGs’ (module) and ‘Targeted genes’ JUN HOMER2 ATF3 VEGFA FOSB NR4A3 MAFF ETS2 MAFF ETS2 KLF10 SESN2 (Purple) JOSD1 ATF3 RARA ATF3 BCOR DDIT4 IER2 GADD45B IER2 GPT2 LONRF3 MIDN HERPUD1 NDNL2 JUNB NR4A2 PHACTR3 DDIT4 ING1 SKP2 FOSL1 RARA AGPAT9 SLC7A1 NR4A2 SIAH2 CDKN1A VEGFA ATF3 BCOR ING1 BHLHE40 METRNL JOSD1 RARA VIT GRIA2 PPP1R15A BHLHE40 CHAC1 PKNOX2 PLCB4 PHF13 SOX9 SYT2 MIDN SOX9 ITPRIP KLF4 BCOR FOS AK5 GADD45B DUSP1 IER2 FOS CDKN1A JUNB STX11 PELO AVPI1 COL7A1 FAM131A TBCCD1 GRIA2 ERLIN1 HERPUD1 SIK1 GPRC5A C1QTNF7 AVPI1 KCNJ15 LATS2 ARC KLF4 BHLHE41 RELT DUSP2 VEGFA FOSB ZC3H12A LMO2 PELO SIK1 LONRF3 SPRY4 ARHGEF2 ARHGEF2 C1QTNF7 TFPI DUSP2 PKNOX2 RGS17 KCNJ15 LPAL2 FOSL1 HK1 NRCAM GPRC5A PPP1R15A CERK DENND3 RARA AGPAT9 DUSP1 CCNA2 SERTAD3 NR4A2 ACVR1B RARA SIAH1 RASSF5 CERK ZNF331 AK5 RELT BRD2 KCTD21 SKP2 NPAS2 ITPRIP NPAS2 SMOX RBM24 MIDN CCDC85C DUSP2 CARS EGR1 SESN2 RASSF5 ASNS FOS FOSB CCNA2 PIP4K2B SPRY4 ANKRD52 BCOR SIAH2 LMO2 DENND3 NR4A3 VIT TNFRSF1B ASTN2 PHACTR3 ASNS FEM1C TNFRSF1B SNORA80B CSRNP3 ITPRIP BNIP3L ATF3 ZNF331 RARA ZC3H12A CHAC1 ASTN2 VEZF1 DUSP1 SNORA80B KLF10 LPAL2 UBE2O EGR1 NR4A2 PCK1 COL7A1 PCK1 STX11 RRAGC PCK1 RBM24 GPT2 HOMER2 METRNL ARC BHLHE41 GARS C15orf41 FOS C1QTNF7 NPAS2 CSRNP3 NRCAM RGS17 VIT RGS17 UBE2O SOX9 KCTD21 NR4A3 RARA SMOX RELT GADD45B ATF3 SERTAD1 RARA BCOR BHLHE40 JUNB TINAGL1 ETS2 KLF10 GADD45B (Purple) LONRF3 COL7A1 TNFRSF1B MMP13 ATF3 SLC2A1 PIM2 CDKN1A ZC3H12A VEZF1 ARC ANKRD52 -

Identification of Genomic Targets of Krüppel-Like Factor 9 in Mouse Hippocampal
Identification of Genomic Targets of Krüppel-like Factor 9 in Mouse Hippocampal Neurons: Evidence for a role in modulating peripheral circadian clocks by Joseph R. Knoedler A dissertation submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy (Neuroscience) in the University of Michigan 2016 Doctoral Committee: Professor Robert J. Denver, Chair Professor Daniel Goldman Professor Diane Robins Professor Audrey Seasholtz Associate Professor Bing Ye ©Joseph R. Knoedler All Rights Reserved 2016 To my parents, who never once questioned my decision to become the other kind of doctor, And to Lucy, who has pushed me to be a better person from day one. ii Acknowledgements I have a huge number of people to thank for having made it to this point, so in no particular order: -I would like to thank my adviser, Dr. Robert J. Denver, for his guidance, encouragement, and patience over the last seven years; his mentorship has been indispensable for my growth as a scientist -I would also like to thank my committee members, Drs. Audrey Seasholtz, Dan Goldman, Diane Robins and Bing Ye, for their constructive feedback and their willingness to meet in a frequently cold, windowless room across campus from where they work -I am hugely indebted to Pia Bagamasbad and Yasuhiro Kyono for teaching me almost everything I know about molecular biology and bioinformatics, and to Arasakumar Subramani for his tireless work during the home stretch to my dissertation -I am grateful for the Neuroscience Program leadership and staff, in particular -

Supplemental Figure Legends
Supplemental Figure Legends Supplemental Figure 1. Expression of cytoplasmic immunoglobulin in D4 cell subsets. A. Phenotype of D4 cell subsets. B. Expression of cyIgM, cyIgA, and cyIgG in D4 cell subsets. Results are those of one experiment representative of three, SI means Staining Index. C. Expression of cyIgM, cyIgA, and cyIgG in D4 cell subsets. Results are mean SI obtained in three separate experiments. * The mean expression is significantly different from that in CD20high B cells. ** The mean expression is significantly different from that in D4 CD20low prePBs. Supplemental Figure 2. Expression of genes coding for B or PC markers. Data are the Affymetrix signals of expression of genes coding for B or PC markers in MBCs, CD20high B cells, CD20low prePBs, CD20- prePBs, PBs, early PCs, and BMPCs using the same color code as in Figure 6A. Data are the mean value ± SD of gene expression determined in 5 separate experiments. * The mean expression is significantly different from that in CD20high B cells. ** The mean expression is significantly different from that in D4 CD20low prePBs. Supplemental Figure 3. Expression of genes coding for homing molecules. Data are the Affymetrix signals of expression of genes coding for remarkable chemokine receptors or integrins in MBCs, CD20high B cells, CD20low prePBs, CD20- prePBs, PBs, early PCs, and BMPCs using the same color code as in Figure 6A. Data are the mean value ± SD of gene expression determined in 5 separate experiments. * The mean expression is significantly different from that in CD20high B cells. ** The mean expression is significantly different from that in D4 CD20low prePBs.