Biochemistry
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-

Coordination of T Cell Activation and Migration Through Formation of the Immunological Synapse
Coordination of T Cell Activation and Migration through Formation of the Immunological Synapse MICHAEL L. DUSTIN Program in Molecular Pathogenesis, Skirball Institute of Biomolecular Medicine and the Department of Pathology, New York University School of Medicine, New York, New York 10016, USA ABSTRACT: T cell activation is based on interactions of T cell antigen receptors with MHC-peptide complexes in a specialized cell–cell junction between the T cell and antigen-presenting cell—the immunological synapse. The immunolog- ical synapse coordinates naïve T cell activation and migration by stopping T cell migration with antigen-presenting cells bearing appropriate major histo- compatibility complex (MHC) peptide complexes. At the same time, the immu- nological synapse allows full T cell activation through sustained signaling over a period of several hours. The immunological synapse supports activation in the absence of continued T cell migration, which is required for T cell activa- tion through serial encounters. Src and Syk family kinases are activated early in immunological synapse formation, but this signaling process returns to the basal level after 30 min; at the same time, the interactions between T cell re- ceptors (TCRs) and MHC peptides are stabilized within the immunological synapse. The molecular pattern of the mature synapse in helper T cells is a self- stabilized structure that is correlated with cytokine production and prolifera- tion. I propose that this molecular pattern and its specific biochemical constit- uents are necessary -

The New Aspects of Aminoacyl-Trna Synthetases
Vol. 47 No. 3/2000 821–834 QUARTERLY Review The new aspects of aminoacyl-tRNA synthetases. Maciej Szymañski, Marzanna Deniziak and Jan Barciszewski½ Institute of Bioorganic Chemistry of the Polish Academy of Sciences, 60-704 Poznañ, Poland Received: 29 December, 1999; revised: 24 May, 2000; accepted: 02 June, 2000 Key words: aminoacylation, aminoacyl-tRNA synthetases, protein biosynthesis, tRNA Aminoacyl-tRNA synthetases (AARS) are essential proteins found in all living organ- isms. They form a diverse group of enzymes that ensure the fidelity of transfer of ge- netic information from the DNA into the protein. AARS catalyse the attachment of amino acids to transfer RNAs and thereby establish the rules of the genetic code by virtue of matching the nucleotide triplet of the anticodon with its cognate amino acid. Here we summarise the effects of recent studies on this interesting family of multifunctional enzymes. The universal genetic code is established in a only components of the gene expression appa- single aminoacylation reaction of transfer ri- ratus that function at the interface between bonucleic acids (tRNAs). The reaction is nucleic acids and proteins. This leads to three catalysed by the family of aminoacyl-tRNA interesting aspects of studies on amino- synthetases (AARS) each of which activates acyl-tRNA synthetases: (i) the mechanism of an amino acid by binding to ATP and trans- amino acid recognition and chemical activa- fers it to the 3¢ end of the cognate tRNA. The tion, (ii) the specificity of tRNA recognition, conservation of the genetic code suggests that and (iii) the origin and evolution of AARS [3]. -

Lymphocyte-Activation Gene 3 (LAG-3) Immune Pathway
Lymphocyte-Activation Gene 3 (LAG-3) About LAG-3 LAG-3 Lymphocyte-activation gene 3 (LAG-3) is an immune checkpoint receptor protein found on the cell surface of effector T cells and regulatory T cells (Tregs) and functions to control T cell response, activation and growth.1 TCR T cells are a type of white blood cell that are part of the immune system. Activation of cytotoxic T cells by antigens enables them to 1 kill unhealthy or foreign cells. Inactive T cell Antigen MHC Dendritic cell (APC) LAG-3 and LAG-3 and LAG-3 and Immune Function T Cell Exhaustion Cancer • After a T cell is activated to kill its • However, in certain situations where T • Because of its critical role in regulating target cell, LAG-3 expression is cells experience prolonged exposure to an exhaustion of cytotoxic T cells and Treg increased to turn off the immune antigen, such as cancer or chronic function, LAG-3 has become a target of response, so that the T cell does not go infection, the T cells become desensitized study in the cancer field. on to attack healthy cells.2 and lose their ability to activate and multiply in the presence of the antigen.4 • In cancer, LAG-3 expressing exhausted • Inhibition of the immune response is cytotoxic T cells and Tregs expressing accomplished through activation of • The desensitized T cell will also LAG-3 gather at tumor sites.5,6 the LAG-3 pathway, which can occur progressively fail to produce cytokines via binding of LAG-3 to a type of (proteins that assist in the immune • Preclinical studies suggest that inhibiting antigen-presenting complex called response) and kill the target cells.4 LAG-3 allows T cells to regain their MHC II. -

An in Vitro Approach to Detect Metabolite Toxicity Due to CYP3A4
Toxicology 216 (2005) 154–167 An in vitro approach to detect metabolite toxicity due to CYP3A4-dependent bioactivation of xenobiotics Luisella Vignati ∗, Elisa Turlizzi 1, Sonia Monaci, Pietro Grossi, Ruben de Kanter, Mario Monshouwer 2 Department of Pre-Clinical Development, Nerviano Medical Sciences S.r.l., V.le Pasteur, 10, 20014, Nerviano, MI, Italy Received 22 June 2005; received in revised form 3 August 2005; accepted 3 August 2005 Available online 19 September 2005 Abstract Many adverse drug reactions are caused by the cytochrome P450 (CYP) dependent activation of drugs into reactive metabolites. In order to reduce attrition due to metabolism-mediated toxicity and to improve safety of drug candidates, we developed two in vitro cell-based assays by combining an activating system (human CYP3A4) with target cells (HepG2 cells): in the first method we incubated microsomes containing cDNA-expressed CYP3A4 together with HepG2 cells; in the second approach HepG2 cells were transiently transfected with CYP3A4. In both assay systems, CYP3A4 catalyzed metabolism was found to be comparable to the high levels reported in hepatocytes. Both assay systems were used to study ten CYP3A4 substrates known for their potential to form metabolites that exhibit higher toxicity than the parent compounds. Several endpoints of toxicity were evaluated, and the measurement of MTT reduction and intracellular ATP levels were selected to assess cell viability. Results demonstrated that both assay systems are capable to metabolize the test compounds leading to increased toxicity, compared to their respective control systems. The co-incubation with the CYP3A4 inhibitor ketoconazole confirmed that the formation of reactive metabolites was CYP3A4 dependent. -

Vaccine Immunology Claire-Anne Siegrist
2 Vaccine Immunology Claire-Anne Siegrist To generate vaccine-mediated protection is a complex chal- non–antigen-specifc responses possibly leading to allergy, lenge. Currently available vaccines have largely been devel- autoimmunity, or even premature death—are being raised. oped empirically, with little or no understanding of how they Certain “off-targets effects” of vaccines have also been recog- activate the immune system. Their early protective effcacy is nized and call for studies to quantify their impact and identify primarily conferred by the induction of antigen-specifc anti- the mechanisms at play. The objective of this chapter is to bodies (Box 2.1). However, there is more to antibody- extract from the complex and rapidly evolving feld of immu- mediated protection than the peak of vaccine-induced nology the main concepts that are useful to better address antibody titers. The quality of such antibodies (e.g., their these important questions. avidity, specifcity, or neutralizing capacity) has been identi- fed as a determining factor in effcacy. Long-term protection HOW DO VACCINES MEDIATE PROTECTION? requires the persistence of vaccine antibodies above protective thresholds and/or the maintenance of immune memory cells Vaccines protect by inducing effector mechanisms (cells or capable of rapid and effective reactivation with subsequent molecules) capable of rapidly controlling replicating patho- microbial exposure. The determinants of immune memory gens or inactivating their toxic components. Vaccine-induced induction, as well as the relative contribution of persisting immune effectors (Table 2.1) are essentially antibodies— antibodies and of immune memory to protection against spe- produced by B lymphocytes—capable of binding specifcally cifc diseases, are essential parameters of long-term vaccine to a toxin or a pathogen.2 Other potential effectors are cyto- effcacy. -

Introduction to Theory of Chemical Reactions
INTRODUCTION TO THEORY OF CHEMICAL REACTIONS BACKGROUND The introductory part of the organic chemistry course has three major modules: Molecular architecture (structure), molecular dynamics (conformational analysis), and molecular transformations (chemical reactions). An understanding of the first two is crucial to an understanding of the third one. The rest of the organic chemistry course will be largely spent on studying different kinds of reactions and mechanisms. A summary of key concepts follows. 1. MOLECULAR ARCHITECTURE - Basic principles of molecular structure. a) Atomic structure b) Orbitals and hybridization c) Covalent bonding d) Lewis structures and resonance forms e) Isomerism, structural and geometric isomers f) Polarity, functional groups, nomenclature systems g) Three-dimensional structures, stereochemistry, stereoisomers. 2. MOLECULAR DYNAMICS - Basic principles of molecular motion involving rotation around single bonds and no bond breakage. The focus is on conformations and their energy relationships, especially in reference to alkanes and cycloalkanes (conformational analysis). a) Steric interactions b) Torsional strain and Newman projections c) Angle strain in cycloalkanes d) Conformations of cyclohexane and terminology associated with it. 3. MOLECULAR TRANSFORMATIONS - This is the part that comprises the bulk of organic chemistry courses. It is the study of chemical reactions and the principles that rule transformations. There are three major aspects of this module. In organic chemistry I we will focus largely on the first two, and leave the study of synthetic strategy for later. a) Reaction mechanisms - Step by step accounts of how electron movement takes place when bonds are broken and formed, and the conditions that favor these processes (driving forces). An understanding of the basic concepts of thermodynamics and kinetics is important here. -
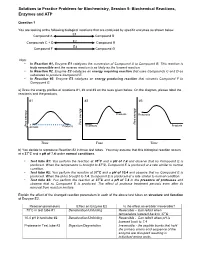
Practice Problems for Biochemistry, Session 5: Biochemical Reactions, Enzymes and ATP
Solutions to Practice Problems for Biochemistry, Session 5: Biochemical Reactions, Enzymes and ATP Question 1 You are looking at the following biological reactions that are catalyzed by specific enzymes as shown below. E1 Compound A Compound B Compounds C + D E2 Compound E E3 Compound F Compound G Note: • In Reaction #1, Enzyme E1 catalyzes the conversion of Compound A to Compound B. This reaction is truly reversible and the reverse reaction is as likely as the forward reaction. • In Reaction #2, Enzyme E2 catalyzes an energy requiring reaction that uses Compounds C and D as substrates to produce Compound E. • In Reaction #3, Enzyme E3 catalyzes an energy producing reaction that converts Compound F to Compound G. a) Draw the energy profiles of reactions #1, #2 and #3 on the axes given below. On the diagram, please label the reactants and the products. #1 #2 #3 y y g g y r r g r Products Reactants ne ne e e ne e e e e e e e Fr Fr Fr Products Reactants Products Reactants Time Time Time b) You decide to reproduce Reaction #2 in three test tubes. You may assume that this biological reaction occurs at a 37oC and a pH of 7.4 under normal conditions. o • Test tube #1: You perform the reaction at 70 C and a pH of 7.4 and observe that no Compound E is produced. When the temperature is brought to 37oC, Compound E is produced at a rate similar to normal condition. o • Test tube #2: You perform the reaction at 37 C and a pH of 10.4 and observe that no Compound E is produced. -

View of Methods for Visceral Adipose Tissue Analysis
Evaluating the role of fibroblast activation protein and fibroblast growth factor 21 in growth hormone-induced adipose tissue fibrosis ____________________________________ A Thesis Presented to The Honors Tutorial College Ohio University _______________________________________ In Partial Fulfillment of the Requirements for Graduation from the Honors Tutorial College with the degree of Bachelor of Science in Biological Sciences ______________________________________ by Delaney Kate Geitgey April 2020 TABLE OF CONTENTS TABLE OF CONTENTS .............................................................................................................. 2 ABSTRACT .................................................................................................................................... 4 INTRODUCTION.......................................................................................................................... 6 Characteristics of adipose tissue and adipocytes ........................................................................ 7 Adipocyte function....................................................................................................................... 7 Adipose tissue depots ................................................................................................................... 8 Main classifications ................................................................................................................. 8 Subcutaneous depots ............................................................................................................... -

Light Guided In-Vivo Activation of Innate Immune Cells with Photocaged TLR 2/6 Agonist
bioRxiv preprint doi: https://doi.org/10.1101/128942; this version posted April 20, 2017. The copyright holder for this preprint (which was not certified by peer review) is the author/funder. All rights reserved. No reuse allowed without permission. Light Guided In-vivo Activation of Innate Immune Cells with Photocaged TLR 2/6 Agonist Keun Ah Ryu†, Bethany McGonnigal†, Troy Moore†, Rock J. Mancini†, Aaron P. Esser-Kahn†,* †Department of Chemistry, University of California, Irvine, Irvine, CA 92697 Abstract The complexity of the immune system creates challenges in exploring its importance and robustness. To date, there have been few techniques developed to manipulate individual components of the immune system in an in vivo environment. Here we show a light-based dendritic cell (DC) activation allowing spatial and temporal control of immune activation in vivo. Additionally, we show time dependent changes in RNA profiles of the draining lymph node, suggesting a change in cell profile following DC migration and indicating that the cells migrating have been activated towards antigen presentation. Introduction Harnessing the innate and adaptive immune response has led to the development of vaccines and therapeutics.1-3 However, as the immune system “rivals the nervous system in complexity,4” understanding how to design better responses and therapies remains a challenge. One area of complexity is the presentation of antigens by the innate system to the adaptive system – including chemical signaling, spatial migration and cell-cell signaling. During this process, dendritic cells, activated by Toll-like receptors (TLRs) convey pathogenic information to the cells of the adaptive immune systems through the production of cytokines and cell surface markers.5, 6 This process involves the migration of activated DCs into lymphatics to present antigens to T-cells.7-10 However, understanding this complex system by manipulating sets of cells within it has been a challenge. -

Inhibition of Microsomal Epoxide Hydrolases by Ureas, Amides, and Amines
Chem. Res. Toxicol. 2001, 14, 409-415 409 Inhibition of Microsomal Epoxide Hydrolases by Ureas, Amides, and Amines Christophe Morisseau, John W. Newman, Deanna L. Dowdy, Marvin H. Goodrow, and Bruce D. Hammock* Department of Entomology and University of California Cancer Center, University of California, Davis, California 95616 Received August 11, 2000 The microsomal epoxide hydrolase (mEH) plays a significant role in the metabolism of xenobiotics such as polyaromatic toxicants. Additionally, polymorphism studies have underlined a potential role of this enzyme in relation to several diseases, such as emphysema, spontaneous abortion, and several forms of cancer. To provide new tools for studying the function of mEH, inhibition of this enzyme was investigated. Inhibition of recombinant rat and human mEH was achieved using primary ureas, amides, and amines. Several of these compounds are more potent than previously published inhibitors. Elaidamide, the most potent inhibitor that is obtained, has a Ki of 70 nM for recombinant rat mEH. This compound interacts with the enzyme forming a noncovalent complex, and blocks substrate turnover through an apparent mix of competitive and noncompetitive inhibition kinetics. Furthermore, in insect cell cultures expressing rat mEH, elaidamide enhances the toxicity effects of epoxide-containing xenobiotics. These inhibitors could be valuable tools for investigating the physiological and toxicological roles of mEH. Introduction Over the past decade, mEH was also described as mediating the transport of bile acid into hepatocytes (14, Epoxides are highly strained three-membered cyclic 15). The mechanism by which mEH participates in bile ethers that are often electrophilically reactive mutagens, absorption is not known. Obtaining potent mEH inhibi- carcinogens, or cytotoxins (1). -

Activation, and Cytokine Production
The Schistosome Granuloma: Characterization of Lymphocyte Migration, Activation, and Cytokine Production This information is current as Catherine A. Rumbley, S. Ali Zekavat, Hiroko Sugaya, Peter of September 27, 2021. J. Perrin, Mohamad Ali Ramadan and S. Michael Phillips J Immunol 1998; 161:4129-4137; ; http://www.jimmunol.org/content/161/8/4129 Downloaded from References This article cites 65 articles, 33 of which you can access for free at: http://www.jimmunol.org/content/161/8/4129.full#ref-list-1 Why The JI? Submit online. http://www.jimmunol.org/ • Rapid Reviews! 30 days* from submission to initial decision • No Triage! Every submission reviewed by practicing scientists • Fast Publication! 4 weeks from acceptance to publication *average by guest on September 27, 2021 Subscription Information about subscribing to The Journal of Immunology is online at: http://jimmunol.org/subscription Permissions Submit copyright permission requests at: http://www.aai.org/About/Publications/JI/copyright.html Email Alerts Receive free email-alerts when new articles cite this article. Sign up at: http://jimmunol.org/alerts The Journal of Immunology is published twice each month by The American Association of Immunologists, Inc., 1451 Rockville Pike, Suite 650, Rockville, MD 20852 Copyright © 1998 by The American Association of Immunologists All rights reserved. Print ISSN: 0022-1767 Online ISSN: 1550-6606. The Schistosome Granuloma: Characterization of Lymphocyte Migration, Activation, and Cytokine Production1 Catherine A. Rumbley, S. Ali Zekavat, Hiroko Sugaya,2 Peter J. Perrin, Mohamad Ali Ramadan, and S. Michael Phillips3 Granuloma formation and its regulation are dependent on lymphocytes. Therefore, we compared the characteristics of lympho- cytes derived from the spleens and granulomas of Schistosoma mansoni-infected mice during the course of their disease. -

Imaging of Fibroblast Activation Protein Alpha Expression in a Preclinical Mouse Model of Glioma Using Positron Emission Tomography
molecules Article Imaging of Fibroblast Activation Protein Alpha Expression in a Preclinical Mouse Model of Glioma Using Positron Emission Tomography Darpan N. Pandya 1, Akesh Sinha 1, Hong Yuan 2, Lysette Mutkus 3 , Kristina Stumpf 3 , Frank C. Marini 3 and Thaddeus J. Wadas 1,* 1 Department of Radiology, University of Iowa, Iowa City, IA 52242, USA; [email protected] (D.N.P.); [email protected] (A.S.) 2 Department of Radiology, University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA; [email protected] 3 Department of Regenerative Medicine, Wake Forest University Health Sciences, Winston-Salem, NC 27157, USA; [email protected] (L.M.); [email protected] (K.S.); [email protected] (F.C.M.) * Correspondence: [email protected]; Tel.: +1-319-335-5009 Academic Editor: Svend Borup Jensen Received: 20 July 2020; Accepted: 11 August 2020; Published: 12 August 2020 Abstract: Glioblastoma multiforme (GBM) is the most aggressive glioma of the primary central nervous system. Due to the lack of effective treatment options, the prognosis for patients remains bleak. Fibroblast activation protein alpha (FAP), a 170 kDa type II transmembrane serine protease was observed to be expressed on glioma cells and within the glioma tumor microenvironment. To understand the utility of targeting FAP in this tumor type, the immuno-PET radiopharmaceutical [89Zr]Zr-Df-Bz-F19 mAb was prepared and Lindmo analysis was used for its in vitro evaluation using the U87MG cell line, which expresses FAP endogenously. Lindmo analysis revealed an association 8 1 constant (Ka) of 10− M− and an immunoreactivity of 52%.