Eschscholzia Californica)
Total Page:16
File Type:pdf, Size:1020Kb
Load more
Recommended publications
-
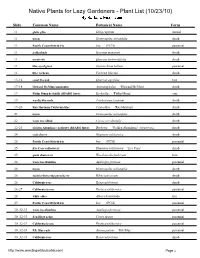
Native Plants for Lazy Gardeners - Plant List (10/23/10)
Native Plants for Lazy Gardeners - Plant List (10/23/10) Slide Common Name Botanical Name Form 11 globe gilia Gilia capitata annual 11 toyon Heteromeles arbutifolia shrub 11 Pacific Coast Hybrid iris Iris (PCH) perennial 11 goldenbush Isocoma menziesii shrub 11 scrub oak Quercus berberidifolia shrub 11 blue-eyed grass Sisyrinchium bellum perennial 11 lilac verbena Verbena lilacina shrub 13-16 coast live oak Quercus agrifolia tree 17-18 Howard McMinn man anita Arctostaphylos 'Howard McMinn' shrub 19 Philip Mun keckiella (RSABG Intro) Keckiella 'Philip Munz' ine 19 woolly bluecurls Trichostema lanatum shrub 19-20 Ray Hartman California lilac Ceanothus 'Ray Hartman' shrub 21 toyon Heteromeles arbutifolia shrub 22 western redbud Cercis occidentalis shrub 22-23 Golden Abundance barberry (RSABG Intro) Berberis 'Golden Abundance' (MAHONIA) shrub 2, coffeeberry Rhamnus californica shrub 25 Pacific Coast Hybrid iris Iris (PCH) perennial 25 Eve Case coffeeberry Rhamnus californica '. e Case' shrub 25 giant chain fern Woodwardia fimbriata fern 26 western columbine Aquilegia formosa perennial 26 toyon Heteromeles arbutifolia shrub 26 fuchsia-flowering gooseberry Ribes speciosum shrub 26 California rose Rosa californica shrub 26-27 California fescue Festuca californica perennial 28 white alder Alnus rhombifolia tree 29 Pacific Coast Hybrid iris Iris (PCH) perennial 30 032-33 western columbine Aquilegia formosa perennial 30 032-33 San Diego sedge Carex spissa perennial 30 032-33 California fescue Festuca californica perennial 30 032-33 Elk Blue rush Juncus patens '.l1 2lue' perennial 30 032-33 California rose Rosa californica shrub http://www weedingwildsuburbia com/ Page 1 30 032-3, toyon Heteromeles arbutifolia shrub 30 032-3, fuchsia-flowering gooseberry Ribes speciosum shrub 30 032-3, Claremont pink-flowering currant (RSA Intro) Ribes sanguineum ar. -

December 2015 / January 2016
University of Arizona Yavapai County Cooperative Extension Yavapai Gardens Master Gardener Newsletter December 2015 - January 2016 Mushrooms for Your Kitchen and Garden By Lori Dekker The world of mushrooms Events & Activities is entering a new era. In the past gardeners and MG Association Meeting, NO MEETING IN DECEM- foodies have considered BER, next meeting Jan. 20 in Prescott. 6:30pm the mushroom to be a Alta Vista Gardening Club, Prescott, fourth Tues- garden novelty or a tasty day of the month, 12:30pm. Call 928-458-9508 for culinary delight, while a information. few of us have an interest in the possible health benefits or the psy- cho/spiritual/recreational uses of a few of the more famous mush- Prescott Area Gourd Society, third Wednesday of the rooms. For now, I’d like to consider the potential health benefits of month, 10:30am, at Miller Valley Indoor Art Market, fungi in your soil and therefore your garden. 531 Madison Ave, Prescott When left to their own devices mushrooms, or more accu- rately fungi, are decomposers and eventually constructors. In a nut- Prescott Orchid Society, 4rd Sunday of the month, 1pm at the Prescott Library, (928) 717-0623 shell, they build soil from the raw material of litter and waste found in the garden. Since they digest food outside their bodies, they are Prescott Area Iris Society call 928-445-8132 for date essentially “sweating” digestive enzymes and producing waste as and place information. they grow through their environment. To put it more simply and hap- pily for gardeners, the fungus breaks down complex compounds Mountain View Garden Club, Prescott Valley, Dewey into more simple ones that then become available, leaving behind area, 2nd Friday of month, 1:30pm, call 775-4993 for metabolites that can, in turn, be utilized by other microbes. -

Landscape Plants Rated by Deer Resistance
E271 Bulletin For a comprehensive list of our publications visit www.rce.rutgers.edu Landscape Plants Rated by Deer Resistance Pedro Perdomo, Morris County Agricultural Agent Peter Nitzsche, Morris County Agricultural Agent David Drake, Ph.D., Extension Specialist in Wildlife Management The following is a list of landscape plants rated according to their resistance to deer damage. The list was compiled with input from nursery and landscape professionals, Cooperative Extension personnel, and Master Gardeners in Northern N.J. Realizing that no plant is deer proof, plants in the Rarely Damaged, and Seldom Rarely Damaged categories would be best for landscapes prone to deer damage. Plants Occasionally Severely Damaged and Frequently Severely Damaged are often preferred by deer and should only be planted with additional protection such as the use of fencing, repellents, etc. Success of any of these plants in the landscape will depend on local deer populations and weather conditions. Latin Name Common Name Latin Name Common Name ANNUALS Petroselinum crispum Parsley Salvia Salvia Rarely Damaged Tagetes patula French Marigold Ageratum houstonianum Ageratum Tropaeolum majus Nasturtium Antirrhinum majus Snapdragon Verbena x hybrida Verbena Brugmansia sp. (Datura) Angel’s Trumpet Zinnia sp. Zinnia Calendula sp. Pot Marigold Catharanthus rosea Annual Vinca Occasionally Severely Damaged Centaurea cineraria Dusty Miller Begonia semperflorens Wax Begonia Cleome sp. Spider Flower Coleus sp. Coleus Consolida ambigua Larkspur Cosmos sp. Cosmos Euphorbia marginata Snow-on-the-Mountain Dahlia sp. Dahlia Helichrysum Strawflower Gerbera jamesonii Gerbera Daisy Heliotropium arborescens Heliotrope Helianthus sp. Sunflower Lobularia maritima Sweet Alyssum Impatiens balsamina Balsam, Touch-Me-Not Matricaria sp. False Camomile Impatiens walleriana Impatiens Myosotis sylvatica Forget-Me-Not Ipomea sp. -

Thesis Draft Rough
Wesleyan University The Honors College Plant-pollinator interactions across California grassland and coastal scrub vegetation types on San Bruno Mountain, San Mateo County by Miles Gordon Brooks Class of 2020 A thesis submitted to the faculty of Wesleyan University in partial fulfillment of the requirements for the Degree of Bachelor of Arts with Departmental Honors from the College of the Environment Middletown, Connecticut April, 2020 1 2 Abstract Animal pollination of plants is a crucial ecosystem service for maintaining biodiversity and ecosystem function, worldwide. High pollinator abundance and diversity can likewise improve the reproductive success of the plant community. Plant-pollinator interaction networks have the potential to identify dominant, specialist, and generalist pollinator species within a system, and their host plant counterparts. Understanding these relationships is paramount for buffering natural systems from biodiversity loss in a world where pollinator abundance continues to decline rapidly. San Bruno Mountain (SBM) in San Mateo County, California, is one of the last natural, open spaces in the urban landscape in the northern San Francisco Peninsula. I conducted a series of timed meanders and vegetation surveys at eight sample sites within SBM (four grassland and four coastal scrub sites) to identify plant species prevalence and pollinator species visitation of flowering plants. I employed a multivariate approach for investigating plant and pollinator species richness, plant and pollinator community composition, and trophic-level interactions across the SBM landscape, and I evaluated differences in these relationships between grassland and coastal scrub habitats. A total of 59 pollinator species and 135 plant species were inventoried over the course of the study. -

Cool Season Annuals
Cool Season Annuals HORT 308/609 Assigned Readings for Plant List 6 Plant List 6 Spring 2020 Read the pages in your textbook associated with the family descriptions and individual taxa covered on Plant List 6 that was distributed in lab. These plant lists are also available on the course website All Text And Images Are Copyrighted By: Dr. Michael A. Arnold, Texas A&M University, Dept. Horticultural Sciences, College Station, TX 77843-2133 Cool season flowers Cool Season A bit of landscaping helps Annuals most any structure! • Tolerant of freezing to subfreezing temperatures – Suitable for use throughout winter in southern half of our region – Suitable for late fall and very early spring use in northern portions of the region • Provides off-season color in winter Cool season foliage Cool (Season) Thoughts Alcea rosea • Many species are derived from edible or Hollyhocks medicinal European species • Classic old-fashioned reseeding annual, • Plants utilized solely for foliage are biennial, or weak perennial more common than with other • Tolerates cold to USDA z. 5, but heat of z. 8 is tough seasonal annuals • Tall cool season annuals are infrequent, • Bold coarse textured foliage; rounded mound the or become tall only late in the season first year or winter and then stiffly upright in spring • Limited range of soil moisture is common • Most decline when day temperatures consistently exceed 80°F or night temperatures exceed 70°F • Mostly for detail designs, bedding, or seasonal containers • Miniature hibiscus-like flowers – Singles quaint, -

Beyond Phenological Onset Dates: Using Observational Data to Detect the Effects of Climate on Collective Properties of Communit
Beyond phenophase onset dates: using observational data to link climate to the collective properties of communities and floras Susan J. Mazer University of California, Santa Barbara Director, California Phenology Project California poppy (Eschscholzia californica) NPN data can now be used to: (1) Compare closely related taxa occupying contrasting climates to examine how phenological sensitivity has diverged. (2) Detect the effects of climatic conditions on community-level phenological metrics (e.g., interspecific variance or synchrony in flowering onset). (3) Identify the climatic parameters that best explain variation in flowering metrics (e.g., flowering onset, duration, termination date, and multiple cycles of flowering) (4) Validate (or question) models constructed using herbarium data. NPN data can be used to: (1) Compare closely related taxa occupying contrasting climates to examine how phenological sensitivity has diverged. 2017. 105: 1610-1622 Western vs. Eastern Oaks • Both groups of oaks respond to warmer/drier conditions by advancing vegetative bud break and/or the onset of flowering • The California oaks are less sensitive to variation in temperature and rainfall across their range • Individual California oak trees have multiple episodes of bud break and flowering each year, while trees of the Eastern oaks exhibit a single event. • Whether these patterns apply to other genera is unknown. NPN data can be used to: (1) Compare closely related taxa occupying contrasting climates to examine how phenological sensitivity has diverged. (2) Detect the effects of climatic conditions on community-level phenological metrics (e.g., interspecific synchrony in flowering onset). (3) Identify the climatic parameters that best explain variation in flowering metrics (4) Validate or question models constructed using herbarium data. -

California Poppy - Eschscholzia Californica
CALIFORNIA POPPY - ESCHSCHOLZIA CALIFORNICA Family: Papaveraceae Part used: immature seed capsules, aerial portions, whole plant Herbal action: sedative, anodyne, anxiolytic, antidepressant Indications: anxiety, nervousness, restless, agitation, insomnia, pain Contraindications and cautions: fever, pregnancy; concurrently with prescription drugs and psychiatric medications. Orally, California poppy can cause muscular stiffness, “morning sluggishness,” and nausea, when used in combination with magnesium and hawthorn. Medicinal uses: Eschscholzia has three important uses in the herbalist’s armamentarium: as a relaxing nervine in anxiety and nervousness; as a sedative in insomnia; and as an anodyne in pain. Eschscholzia tincture in anxiety and nervousness, where there are “…skin hypersensitivities and peripatetic movements”. As an anxiolytic, California Poppy should be taken in smaller doses, combined with herbs such as Pulsatilla (Anenome pulsatilla). When used in higher doses California Poppy acts as a sedative, inducing a pleasant drowsy feeling, not enough to promote marked sedation, but powerful enough that tasks such as driving or operating machinery are best avoided under its influence. In states of pain, such as intestinal colic, rheumatism, toothaches and earaches, California Poppy can be dosed in higher amounts. A more recent usage for Eschscholzia is in the treatment of heroin addiction and withdrawal. Donna Odierna, herbalist and director of the H.E.A.L.T.H. Needle Exchange clinic in Oakland, California, uses Eschscholzia as the primary ingredient in her clinic’s “Kick Juice,” along with smaller amounts of Vitex agnus-castus, Avena sativa, Piper methysticum and Verbena officinalis. In her practice with heroin and methadone addicted patients, Odierna has found this formula helpful to both wean patients off of opioids, as well as to reduce the frequency and amount of heroin or methadone used. -

A Crop of Wildflowers Have Been Seen Popping up in a Variety of Sites Around Southern California, and in Very Accessibl
FINALLY! A crop of wildflowers have been seen popping up in a variety of sites around Southern California, and in very accessible areas, too, if you can take a short holiday escape during spring break. Let’s start along the coastal regions. A wonderful wildflower outing is to be had if you check out the Colorado Lagoon in Long Beach off of 4th Street and Park Ave. Look for wild hyacinth (Dichelostemma capitatum), tidy tips (Layia platyglossa), deerweed (Acmispon glaber), arroyo lupine (Lupinus succulentus), California poppy (Eschscholzia californica), California bush sunflower (Encelia californica), giant coreopsis (Leptosyne sp.), Santa Barbara milkvetch (Astragalus trichopodus), Chinese houses (Collinsia heterophylla), mulefat (Baccharis salicifolia), California buckwheat (Eriogonum fasciculatum), California four o’clock (Mirabilis laevis var. crassifolia) and golden bush (Isocoma sp.). Enjoy walking the new paths around the entire lagoon and marvel at the habitat restoration of this important wetland area! Chinese houses (Collinsia heterophylla) and tidy tips (Layia platyglossa). Photos by George Nanoski. Mostly perennials and a few annual species of flowers can be seen along the Brightwater Trail in the Bolsa Chica Ecological Reserve, and specifically around the ‘pocket’ in Huntington Beach for those who know the reserve. Look for California four o’clock (Mirabilis laevis var. crassifolia), bush sunflower (Encelia californica) and bladderpod (Peritoma arborea). Blue-eye grass (Sisyrinchium bellum) and fiddleneck (Amsinckia sp.) are in sunny open areas along the trails too. By the way, this is a great birding location, so bring the binoculars as well as a wildflower guide. While enjoying fresh ocean breezes and sunshine, there are many showy natives to enjoy along the trails at the Environmental Nature Center in Newport Beach. -

Eschscholzia Californica Cham., CALIFORNIA POPPY. Perennial
Eschscholzia californica Cham., CALIFORNIA POPPY. Perennial herb or annual, taprooted, rosetted, several−many-stemmed at base, spreading to erect, in range 15−60 cm tall; shoots with basal leaves and well-developed cauline leaves, glabrous, glaucous (not glaucous); latex colorless. Stems: 5−8-ridged, to 6 mm diameter, with ridge descending from each cauline leaf, often rose-striped with whitish ridges; hollow. Leaves: helically alternate, deeply 2−4× pinnately dissected with paired, subopposite to opposite lateral subdivisions, long-petiolate, without stipules; petiole semicircular to lenticular in ×-section, 35−110 mm long; blade broadly ovate to ± deltate in outline, to 80 mm long, << petiole, with lobes of each subdivision not arising from same point, terminal portion unequally 3-lobed or 3- divided with each subdivision wedge-shaped; ultimate segments narrowly oblong to narrowly oblanceolate, typically to 5(−13) mm × 0.6−2.9 mm, strawberry pink beneath surface wax especially on tips, obtuse to acute at tip, pinnately veined with principal veins somewhat raised on lower surface, lower surface with blisterlike cells along principal veins and tips. Inflorescence: cyme, terminal, 1–several-flowered, bracteate, glabrous, glaucous; bract subtending cyme leaflike and 2−3× dissected but with shorter petiole than leaf; pedicel erect, inconspicuously angled, to 150 mm long, to 1.3 mm diameter, hollow. Flower: bisexual, radial, 20–60 mm wide, dish-shaped; bud erect; hypanthiumlike receptacle surrounding ovary base, deep cuplike to broadly funnel-shaped -

Project Budburst Available Species Sheet
Project BudBurst Available Species Sheet www.budburst.org Wildflowers and Herbs Deciduous Trees and Shrubs • Alfalfa (Medicago sativa) • American linden (Tilia americana) • American pasqueflower (Pulsatilla patens aka • Antelope bitterbrush (Purshia tridentata) Anemone patens) • Apple (Malus pumila) • Bigleaf lupine (Lupinus polyphyllus) • Bald cypress (Taxodium distichum) • Bitter root (Lewisia rediviva) • Balsam poplar (Populus balsamifera (aka • California poppy (Eschscholzia californica) trichocarpa)) • Canada thistle (Cirsium arvense) • Beaked hazelnut (Corylus cornuta) • Colorado blue columbine (Aquilegia caerulea) • Bigleaf maple (Acer macrophyllum) • Common dandelion (Taraxacum officinale) • Black elderberry (Sambucus nigra) • Common yarrow (Achillea millefolium) • Black locust (Robinia pseudoacacia) • Darkthroat shootingstar (Dodecatheon • Boxelder (Acer negundo) pulchellum) • Chokecherry (Prunus virginiana) • Dogtooth violet (Erythronium americanum) • Common lilac (Syringa vulgaris) • Field mustard (Brassica rapa) • Common snowberry (Symphoricarpos albus) • Henbit deadnettle (Lamium amplexicaule) • Eastern serviceberry (Amelanchier canadensis) • Indian pink (Spigelia marilandica) • Flowering dogwood (Cornus florida) • Jack in the pulpit (Arisaema triphyllum) • Forsythia (Forsythia xintermedia) • Lanceleaf springbeauty (Claytonia lanceolata) • Lewis' mock orange (Philadelphus lewisii) • Large flowered trillium (Trillium grandiflorum) • Pacific dogwood (Cornus nuttallii) • Mayapple (Podophyllum peltatum) • Paper birch (Betula -

Annuals for Nebraska Landscapes I. Flowering Plants
G1774 Annuals for Nebraska Landscapes I. Flowering Plants Dale T. Lindgren, Extension Horticulturist; Anne M. Streich, Extension Horticulture Educator; Kim A. Todd, Extension Landscape Horticulture Specialist; and Steven N. Rodie, Extension Landscape Horticulture Specialist typically required for healthy establishment and growth, are This NebGuide describes those annual flowers that potential disadvantages of using annual flowers. can be grown in Nebraska for use by the home gardener Annuals can be used in beds, borders, rock gardens, in landscape design. window boxes, hanging baskets and container gardens. They can add focal color to entrances or enhance monotonous Annuals are non-woody plants that complete their life landscapes. Annuals can be used in newly planted perennial cycle in one growing season, ending with seed production. or shrub beds and borders, filling the vacant spaces with color Annuals provide color from early summer until frost, an ad- until the permanent plantings mature. vantage over most perennial flowers, which usually have a Consider site conditions and the preferred growing shorter blooming season. Certain annuals, such as moss rose, requirements of annuals when designing with them. Annuals California poppy and alyssum, may self-seed, but most an- are generally incorporated into a design for their color, but nuals must be planted yearly. Some perennial plants that live texture, form, size and mass characteristics also contribute to the from year to year in warmer climates, such as begonias and overall effect. Evaluate the planting site’s sun exposure, wind snapdragons, are included with annuals in Nebraska since they patterns, water requirements, soil type and fertility, and then are not winter-hardy and must be replanted each year. -

California Native Plants Present at the Humboldt Botanical Gardens Lost Coast Brewery Native Plant Garden
California Native Plants Present at the Humboldt Botanical Gardens Lost Coast Brewery Native Plant Garden Abies grandis (grand fir) Acer circinatum (vine maple) Achillea millefolium (common yarrow) Adiantum aleuticum (five finger fern) Aesculus californica (buckeye) Allium unifolium (one leaf onion) Amelanchier alnifolia (western serviceberry) Alnus rubra (red alder) Artemisia californica ‘Canyon Gray’ (Canyon Gray sagebrush) Artemisia californica ‘Montara’ (Montara coastal sagebrush) Artemisia pycnocephala ‘David’s Choice’ (sand hill sage) Arctostaphylos bakeri ‘Louis Edmunds’ (Louis Edmunds manzanita) Arctostaphylos columbiana (hairy manzanita) Arctostaphylos columbiana x media (hybrid manzanita) Arctostaphylos densiflora ‘Howard McMinn’ (Howard McMinn manzanita) Arctostaphylos densiflora 'Sentinel' x Arctostaphylos manzanita 'Dr. Hurd' (Austin Griffiths’ manzanita) Arctostaphylos densiflora ‘Sentinel’ (Sentinel manzanita) Arctostaphylos densiflora ‘White Lanterns’ (White lanterns manzanita) Arctostaphylos franciscana ‘Doyle Drive’ (Franciscan manzanita) Arctostaphylos franciscana ‘Tilden 47 002’ (Franciscan manzanita) Arctostaphylos franciscana ‘Lester Rowntree’ (Franciscan manzanita) Arctostaphylos hookeri x pajaroensis ‘Sunset’ (Sunset manzanita) Arctostaphylos ‘John Dourley’ Arctostaphylos pajaroensis ‘Myrtle Wolf’’ (Myrtle Wolf manzanita) Arctostaphylos pajaroensis ‘Paradise’ (Paradise manzanita) Arctostaphylos pajaroensis ‘Warren Roberts’ (Warren Roberts manzanita) Arctostaphylos manzanita ‘Dr. Hurd’ (Dr. Hurd manzanita) Arctostaphylos